
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,977,066 – 14,977,336 |
| Length | 270 |
| Max. P | 0.993164 |

| Location | 14,977,066 – 14,977,185 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.31 |
| Mean single sequence MFE | -20.16 |
| Consensus MFE | -16.49 |
| Energy contribution | -16.30 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14977066 119 - 20766785 CGACACACCCUAUGAAAAUCGUAUGCAAUCUGCAGGUAAGUGCAGGUGAUUCAUACACCCACACAUGCACACUUACUCA-AAAAAACUGGUAGAAACCACACAUAUUUCUCGAAAAACAU (((...(((.((((.....))))(((.....))))))..(((((.(((...........)))...))))).........-.......((((....))))..........)))........ ( -18.80) >DroSim_CAF1 7608 120 - 1 CGACACACCCUAUUAAAAUCGUAUGCAAUCUGCAGGUAAGUGCAGGUGAUUCAUACCCCAUCACACGCACACUUACUCAGAAAAAACUGGUAGAAACCACACAUAUUUCUCGCCAAACAU ....................((......((((..((((((((..((((((.........))))).)...))))))))))))......(((((((((.........))))).)))).)).. ( -22.20) >DroEre_CAF1 7399 119 - 1 CGACACACCCAAUAAAAAUCGUGUGCAAUCUGCAGGUAAGUGCAGGUGAUUCAUACCCACACACACAUACACUUACUCA-AAAAAACUGGUAGAAACCACACAUAUUUCUCGCCAAACAU ....................(((((...(((((.((((((((...(((...............)))...))))))))((-.......)))))))...))))).................. ( -21.46) >DroYak_CAF1 7072 118 - 1 CGACACACCCAAUAAAAAUCGUAUGCAAUCUGCAGGUAAGUGCAGGUGAUUCAUACCCACACACACAUACACUUACUCA-AA-AAACUGGUAGAAACCACACAUAUUUCUCGCCAAACAU .......................(((.....)))((((((((...(((...............)))...))))))))..-..-....(((((((((.........))))).))))..... ( -18.16) >consensus CGACACACCCAAUAAAAAUCGUAUGCAAUCUGCAGGUAAGUGCAGGUGAUUCAUACCCACACACACACACACUUACUCA_AAAAAACUGGUAGAAACCACACAUAUUUCUCGCCAAACAU (((.................(((((..(((((((......)))))))....)))))...............................((((....))))..........)))........ (-16.49 = -16.30 + -0.19)

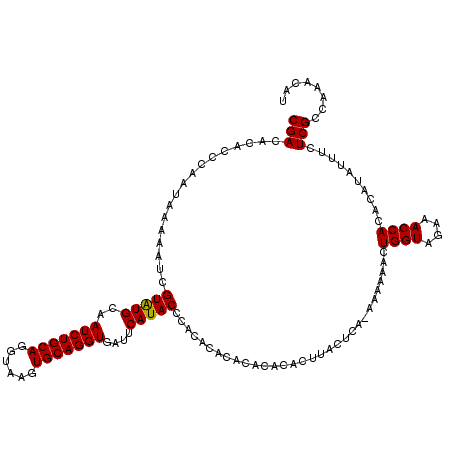
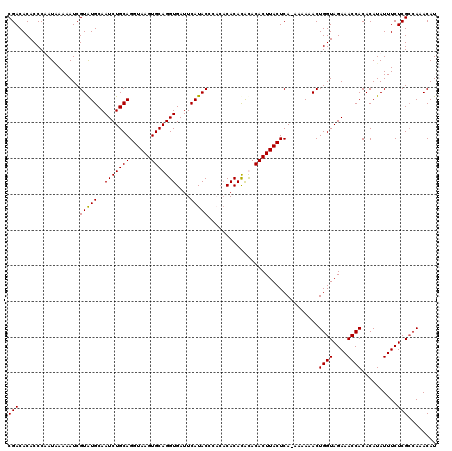
| Location | 14,977,145 – 14,977,241 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 85.13 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -24.15 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14977145 96 + 20766785 CUUACCUGCAGAUUGCAUACGAUUUUCAUAGGGUGUGUCGUUGUUGUCUUGGAGUAGCUACAGCAA-------------CA-ACAACAACAACGACCUUCCCAGACGCAG .....((((((((((....)))))).....(((...(((((((((((..((..((.((....)).)-------------).-.)))))))))))))...)))....)))) ( -32.40) >DroSim_CAF1 7688 96 + 1 CUUACCUGCAGAUUGCAUACGAUUUUAAUAGGGUGUGUCGUUGUUGUCUUGGAGUAGCUACAGCAA-------------CA-ACAACAACAACGACCUUCCCAGACGCAG .....((((((((((....)))))).....(((...(((((((((((..((..((.((....)).)-------------).-.)))))))))))))...)))....)))) ( -32.40) >DroEre_CAF1 7478 105 + 1 CUUACCUGCAGAUUGCACACGAUUUUUAUUGGGUGUGUCGUUGCUGUCUUGGAGUAGCUACA----GCAGAAGCAACAACAC-CAACAACAACGACCUUCCCAGACGCAG .....((((((((((....))))))...(((((...(((((((.(((..((..((.(((...----.....))).))..)).-..))).)))))))...)))))..)))) ( -30.60) >consensus CUUACCUGCAGAUUGCAUACGAUUUUAAUAGGGUGUGUCGUUGUUGUCUUGGAGUAGCUACAGCAA_____________CA_ACAACAACAACGACCUUCCCAGACGCAG .....((((((((((....)))))).....(((...(((((((((((..((................................)))))))))))))...)))....)))) (-24.15 = -24.48 + 0.33)

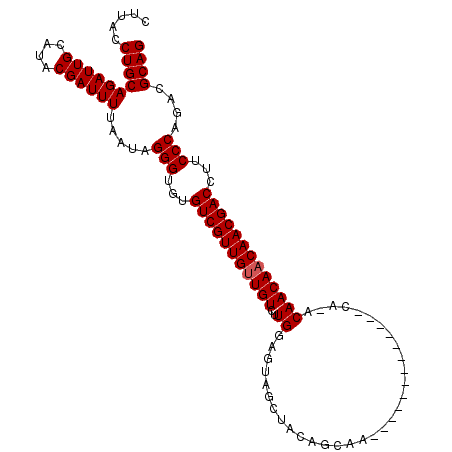
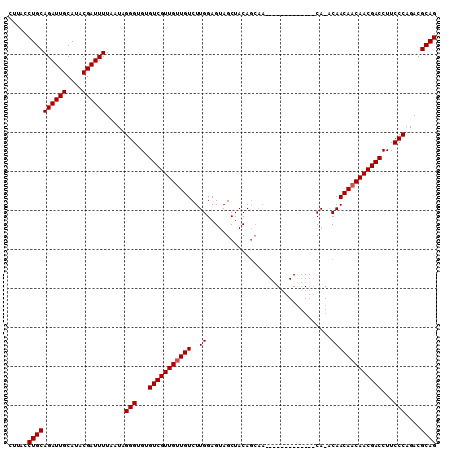
| Location | 14,977,145 – 14,977,241 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 85.13 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -25.71 |
| Energy contribution | -25.49 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14977145 96 - 20766785 CUGCGUCUGGGAAGGUCGUUGUUGUUGU-UG-------------UUGCUGUAGCUACUCCAAGACAACAACGACACACCCUAUGAAAAUCGUAUGCAAUCUGCAGGUAAG ((((....(((...((((((((((((..-.(-------------(.((....)).)).....))))))))))))...)))((((.....))))........))))..... ( -33.00) >DroSim_CAF1 7688 96 - 1 CUGCGUCUGGGAAGGUCGUUGUUGUUGU-UG-------------UUGCUGUAGCUACUCCAAGACAACAACGACACACCCUAUUAAAAUCGUAUGCAAUCUGCAGGUAAG ((((....(((...((((((((((((..-.(-------------(.((....)).)).....))))))))))))...)))..........(....).....))))..... ( -30.80) >DroEre_CAF1 7478 105 - 1 CUGCGUCUGGGAAGGUCGUUGUUGUUG-GUGUUGUUGCUUCUGC----UGUAGCUACUCCAAGACAGCAACGACACACCCAAUAAAAAUCGUGUGCAAUCUGCAGGUAAG ((((...((((...(((((((((((((-(.((.(((((......----.))))).)).)))..)))))))))))...)))).........(....).....))))..... ( -38.10) >consensus CUGCGUCUGGGAAGGUCGUUGUUGUUGU_UG_____________UUGCUGUAGCUACUCCAAGACAACAACGACACACCCUAUAAAAAUCGUAUGCAAUCUGCAGGUAAG ((((....(((...((((((((((((....................................))))))))))))...)))..........(....).....))))..... (-25.71 = -25.49 + -0.22)

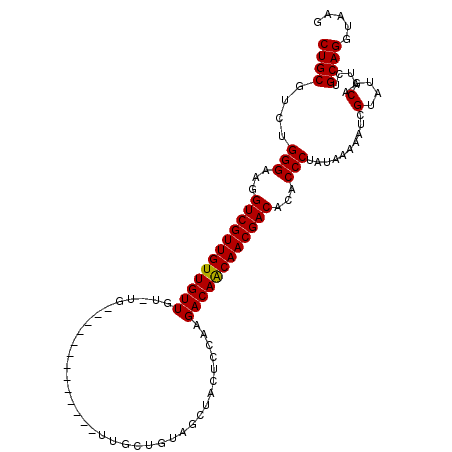
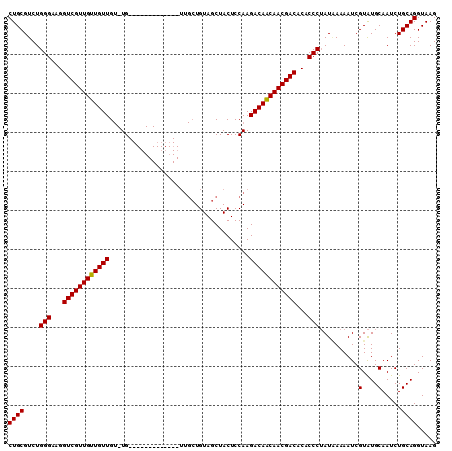
| Location | 14,977,211 – 14,977,315 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 97.62 |
| Mean single sequence MFE | -21.89 |
| Consensus MFE | -20.84 |
| Energy contribution | -20.85 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14977211 104 + 20766785 -CA-ACAACAACAACGACCUUCCCAGACGCAGAACUUUGGCAAAAAUGCCAAGCUCGCUCGCUCGCUCGCCACAUCCACAGAAUUACAAUCCACAGUGGCGGCAGC -..-.....................((.((.((.(((.((((....))))))).))))))(((.((.((((((......................))))))))))) ( -25.25) >DroSec_CAF1 11 104 + 1 -CA-ACAACAACAACGACCUUCCCAGACGCAGAACUUUGGCAAAAAUGCCAAGCUCGCUCGCUCGCUCGCCACAUCCACAGAAUUACAAUCCACAUCGGCGGCAGC -..-.....................((.((.((.(((.((((....))))))).))))))(((.((.((((..........................))))))))) ( -20.77) >DroSim_CAF1 7754 104 + 1 -CA-ACAACAACAACGACCUUCCCAGACGCAGAACUUUGGCAAAAAUGCCAAGCUCGCUCGCUCGCUCGCCACAUCCACAGAAUUACAAUCCACAUCGGCGGCAGC -..-.....................((.((.((.(((.((((....))))))).))))))(((.((.((((..........................))))))))) ( -20.77) >DroEre_CAF1 7552 105 + 1 ACAC-CAACAACAACGACCUUCCCAGACGCAGAACUUUGGCAAAAAUGCCAAGCUCGCUCGCUCGCUCGCCACAUCCACAGAAUUACAAUCCACAUCGGCGGCAGC ....-....................((.((.((.(((.((((....))))))).))))))(((.((.((((..........................))))))))) ( -20.77) >consensus _CA_ACAACAACAACGACCUUCCCAGACGCAGAACUUUGGCAAAAAUGCCAAGCUCGCUCGCUCGCUCGCCACAUCCACAGAAUUACAAUCCACAUCGGCGGCAGC .........................((.((.((.(((.((((....))))))).))))))(((.((.((((..........................))))))))) (-20.84 = -20.85 + 0.00)

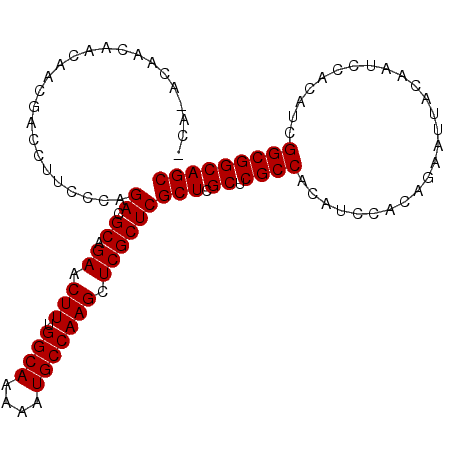
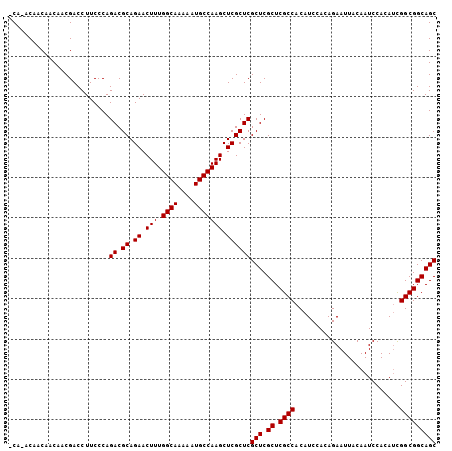
| Location | 14,977,241 – 14,977,336 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 99.16 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -23.83 |
| Energy contribution | -23.83 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14977241 95 + 20766785 AACUUUGGCAAAAAUGCCAAGCUCGCUCGCUCGCUCGCCACAUCCACAGAAUUACAAUCCACAGUGGCGGCAGCCACAAUGGCAAAAGUAGCCAG ...(((((((....)))))))((.(((.(((.((.((((((......................)))))))).(((.....)))...)))))).)) ( -28.25) >DroSec_CAF1 41 95 + 1 AACUUUGGCAAAAAUGCCAAGCUCGCUCGCUCGCUCGCCACAUCCACAGAAUUACAAUCCACAUCGGCGGCAGCCACAAUGGCAAAAGUAGCCAG ...(((((((....)))))))((.(((.(((.((.((((..........................)))))).(((.....)))...)))))).)) ( -23.77) >DroSim_CAF1 7784 95 + 1 AACUUUGGCAAAAAUGCCAAGCUCGCUCGCUCGCUCGCCACAUCCACAGAAUUACAAUCCACAUCGGCGGCAGCCACAAUGGCAAAAGUAGCCAG ...(((((((....)))))))((.(((.(((.((.((((..........................)))))).(((.....)))...)))))).)) ( -23.77) >DroEre_CAF1 7583 95 + 1 AACUUUGGCAAAAAUGCCAAGCUCGCUCGCUCGCUCGCCACAUCCACAGAAUUACAAUCCACAUCGGCGGCAGCCACAAUGGCAAAAGUAGCCAG ...(((((((....)))))))((.(((.(((.((.((((..........................)))))).(((.....)))...)))))).)) ( -23.77) >DroYak_CAF1 7270 95 + 1 AACUUUGGCAAAAAUGCCAAGCUCGCUCGCUCGCUCGCCACAUCCACAGAAUUACAAUCCACAUCGGCGGCAGCCACAAUGGCAAAAGUAGCCAG ...(((((((....)))))))((.(((.(((.((.((((..........................)))))).(((.....)))...)))))).)) ( -23.77) >consensus AACUUUGGCAAAAAUGCCAAGCUCGCUCGCUCGCUCGCCACAUCCACAGAAUUACAAUCCACAUCGGCGGCAGCCACAAUGGCAAAAGUAGCCAG ...(((((((....)))))))((.(((.(((.((.((((..........................)))))).(((.....)))...)))))).)) (-23.83 = -23.83 + 0.00)

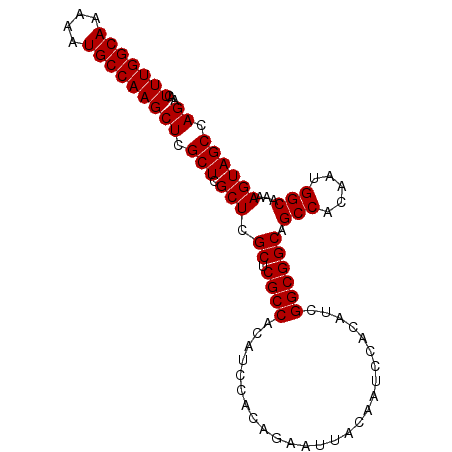
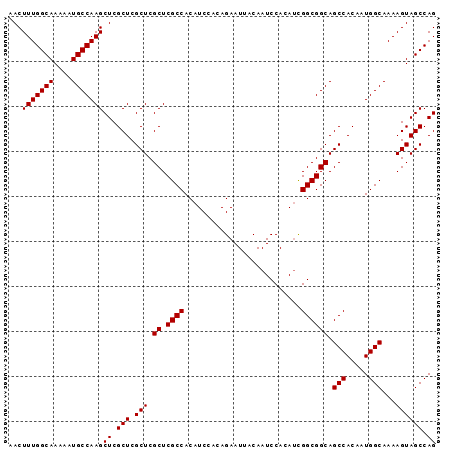
| Location | 14,977,241 – 14,977,336 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 99.16 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -32.26 |
| Energy contribution | -32.10 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14977241 95 - 20766785 CUGGCUACUUUUGCCAUUGUGGCUGCCGCCACUGUGGAUUGUAAUUCUGUGGAUGUGGCGAGCGAGCGAGCGAGCUUGGCAUUUUUGCCAAAGUU .((((.......))))((((.(((((((((((.(..((.......))..)....)))))).)).)))..))))((((((((....)))).)))). ( -34.80) >DroSec_CAF1 41 95 - 1 CUGGCUACUUUUGCCAUUGUGGCUGCCGCCGAUGUGGAUUGUAAUUCUGUGGAUGUGGCGAGCGAGCGAGCGAGCUUGGCAUUUUUGCCAAAGUU .((((.......))))((((.((((((((((.((..((.......))..))....))))).)).)))..))))((((((((....)))).)))). ( -32.40) >DroSim_CAF1 7784 95 - 1 CUGGCUACUUUUGCCAUUGUGGCUGCCGCCGAUGUGGAUUGUAAUUCUGUGGAUGUGGCGAGCGAGCGAGCGAGCUUGGCAUUUUUGCCAAAGUU .((((.......))))((((.((((((((((.((..((.......))..))....))))).)).)))..))))((((((((....)))).)))). ( -32.40) >DroEre_CAF1 7583 95 - 1 CUGGCUACUUUUGCCAUUGUGGCUGCCGCCGAUGUGGAUUGUAAUUCUGUGGAUGUGGCGAGCGAGCGAGCGAGCUUGGCAUUUUUGCCAAAGUU .((((.......))))((((.((((((((((.((..((.......))..))....))))).)).)))..))))((((((((....)))).)))). ( -32.40) >DroYak_CAF1 7270 95 - 1 CUGGCUACUUUUGCCAUUGUGGCUGCCGCCGAUGUGGAUUGUAAUUCUGUGGAUGUGGCGAGCGAGCGAGCGAGCUUGGCAUUUUUGCCAAAGUU .((((.......))))((((.((((((((((.((..((.......))..))....))))).)).)))..))))((((((((....)))).)))). ( -32.40) >consensus CUGGCUACUUUUGCCAUUGUGGCUGCCGCCGAUGUGGAUUGUAAUUCUGUGGAUGUGGCGAGCGAGCGAGCGAGCUUGGCAUUUUUGCCAAAGUU .((((.......))))((((.((((((((((.((..((.......))..))....))))).)).)))..))))((((((((....)))).)))). (-32.26 = -32.10 + -0.16)

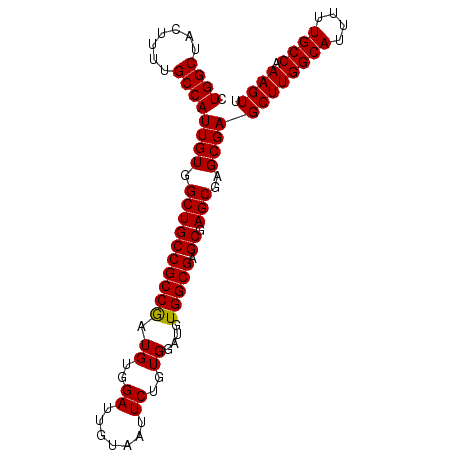
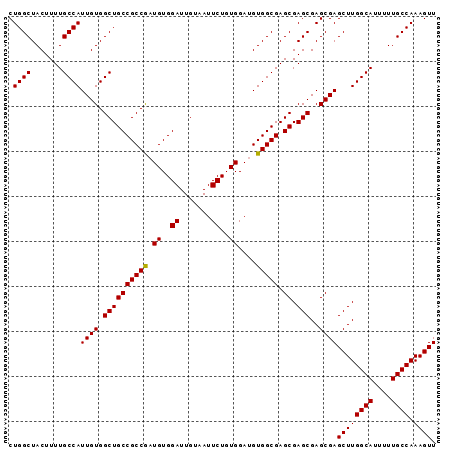
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:17 2006