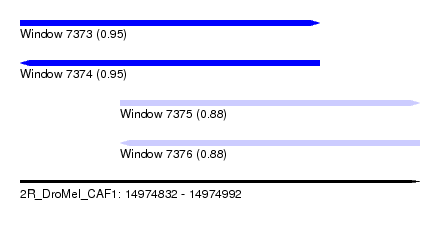
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,974,832 – 14,974,992 |
| Length | 160 |
| Max. P | 0.949459 |

| Location | 14,974,832 – 14,974,952 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -50.95 |
| Consensus MFE | -47.04 |
| Energy contribution | -47.10 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14974832 120 + 20766785 CUGAGGCCCUGCCAGCGGAUAGCGUGUCCAAUGGCACAGCAUCCAAUGUAGCAGCACCGGCGGCGCCAGUAUCGAGCGCAACAAACGCGGUGCCACCACUGGCCGCCGUCUCCAGCACAA (((.((..((((((..((((.(((((((....))))).))))))..))..))))..))((((((((((((...(.((((..(....)..)))))...))))).)))))))..)))..... ( -49.00) >DroSim_CAF1 5515 120 + 1 CUGAGGCCCUGCCAGCGGAUAGCGUGUCCAAUGGCACAGCAUCCAAUGUUGCAGCACCGGCGGCGCCAGUAUCGAGCGCCACAAACGCGGUGCCACCACUGGCCGCCGUCUCCAGCACAA (((.((..((((((..((((.(((((((....))))).))))))..))..))))..))((((((((((((...(.(((((.(....).))))))...))))).)))))))..)))..... ( -52.00) >DroEre_CAF1 5169 120 + 1 CUGAAGCCCUGCCAGCGGAUAGCGUGUCCAAUGGCACAGCAUCCAAUGUUGCAGCACCGGCGGCGCCAGUAUCGAGCGCCACAAAUGCGGUGCCACCACUGGCCGCCGUCUCUAGCACAA .....((.((((((..((((.(((((((....))))).))))))..))..))))....((((((((((((...(.(((((.(....).))))))...))))).)))))))....)).... ( -49.10) >DroYak_CAF1 4831 120 + 1 CUGAAGCCCUGCCAGCGGAUGGUGUGUCCAAUGGCACAGCAUCCAAUGUUGCAGCACCGGCGGCGCCAGUAUCGAGCGCCACAAACGCGGUGCCACCACUGGCCGCCGUCUCCAGCACAA .....((.((((((..(((((.((((((....)))))).)))))..))..))))....((((((((((((...(.(((((.(....).))))))...))))).)))))))....)).... ( -53.70) >consensus CUGAAGCCCUGCCAGCGGAUAGCGUGUCCAAUGGCACAGCAUCCAAUGUUGCAGCACCGGCGGCGCCAGUAUCGAGCGCCACAAACGCGGUGCCACCACUGGCCGCCGUCUCCAGCACAA .....((.((((((..((((.(((((((....))))).))))))..))..))))....((((((((((((...(.(((((.(....).))))))...))))).)))))))....)).... (-47.04 = -47.10 + 0.06)

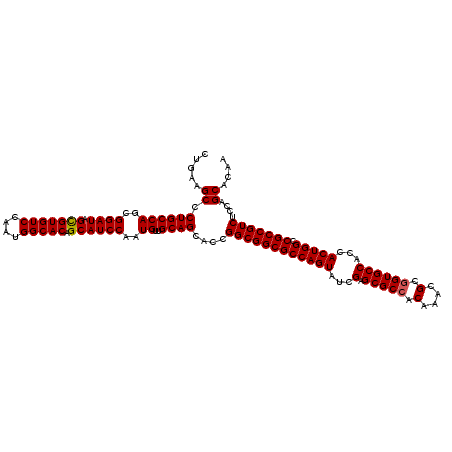
| Location | 14,974,832 – 14,974,952 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -58.38 |
| Consensus MFE | -57.03 |
| Energy contribution | -57.52 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14974832 120 - 20766785 UUGUGCUGGAGACGGCGGCCAGUGGUGGCACCGCGUUUGUUGCGCUCGAUACUGGCGCCGCCGGUGCUGCUACAUUGGAUGCUGUGCCAUUGGACACGCUAUCCGCUGGCAGGGCCUCAG ....((((....))))((((.((((..((((((((......(((((.......))))))).))))))..))))...........(((((.((((.......)))).))))).)))).... ( -56.00) >DroSim_CAF1 5515 120 - 1 UUGUGCUGGAGACGGCGGCCAGUGGUGGCACCGCGUUUGUGGCGCUCGAUACUGGCGCCGCCGGUGCUGCAACAUUGGAUGCUGUGCCAUUGGACACGCUAUCCGCUGGCAGGGCCUCAG ...(((((....))))).(((((((..((((((.....((((((((.......))))))))))))))..)..))))))..(((.(((((.((((.......)))).))))).)))..... ( -60.20) >DroEre_CAF1 5169 120 - 1 UUGUGCUAGAGACGGCGGCCAGUGGUGGCACCGCAUUUGUGGCGCUCGAUACUGGCGCCGCCGGUGCUGCAACAUUGGAUGCUGUGCCAUUGGACACGCUAUCCGCUGGCAGGGCUUCAG .(((((.((...((((((((((((..(((.((((....)))).)))...)))))..)))))))...)))).))).((((.(((.(((((.((((.......)))).))))).))))))). ( -56.50) >DroYak_CAF1 4831 120 - 1 UUGUGCUGGAGACGGCGGCCAGUGGUGGCACCGCGUUUGUGGCGCUCGAUACUGGCGCCGCCGGUGCUGCAACAUUGGAUGCUGUGCCAUUGGACACACCAUCCGCUGGCAGGGCUUCAG ...(((((....)))))((((((.(..((((((.....((((((((.......))))))))))))))..)......(((((.((((((...)).)))).))))))))))).......... ( -60.80) >consensus UUGUGCUGGAGACGGCGGCCAGUGGUGGCACCGCGUUUGUGGCGCUCGAUACUGGCGCCGCCGGUGCUGCAACAUUGGAUGCUGUGCCAUUGGACACGCUAUCCGCUGGCAGGGCCUCAG ...(((((....))))).(((((((..((((((.....((((((((.......))))))))))))))..)..))))))..(((.(((((.((((.......)))).))))).)))..... (-57.03 = -57.52 + 0.50)

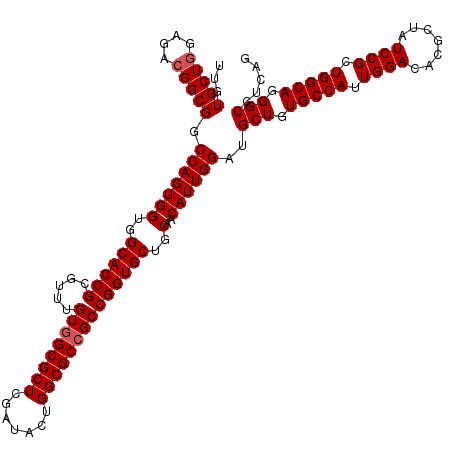
| Location | 14,974,872 – 14,974,992 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -32.15 |
| Energy contribution | -32.65 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14974872 120 + 20766785 AUCCAAUGUAGCAGCACCGGCGGCGCCAGUAUCGAGCGCAACAAACGCGGUGCCACCACUGGCCGCCGUCUCCAGCACAACCGCCACCUACGCCACCAACUCGAUCAGCACAUCCUCGCA .......((((.......((((((((((((...(.((((..(....)..)))))...))))).)))))))....((......))...))))................((........)). ( -32.10) >DroSim_CAF1 5555 120 + 1 AUCCAAUGUUGCAGCACCGGCGGCGCCAGUAUCGAGCGCCACAAACGCGGUGCCACCACUGGCCGCCGUCUCCAGCACAACCGCCACCUACGCCACCAACUCGAUCAGCACAUCCUCGCA .......((((..((...((((((((((((...(.(((((.(....).))))))...))))).)))))))....((......)).......))...)))).......((........)). ( -36.60) >DroEre_CAF1 5209 120 + 1 AUCCAAUGUUGCAGCACCGGCGGCGCCAGUAUCGAGCGCCACAAAUGCGGUGCCACCACUGGCCGCCGUCUCUAGCACAACCGCCACCUACGCCACCAACUCGAUUAGCACAUCCUCGCA .......((((..((...((((((((((((...(.(((((.(....).))))))...))))).)))))))....((......)).......))...)))).......((........)). ( -36.90) >DroYak_CAF1 4871 111 + 1 AUCCAAUGUUGCAGCACCGGCGGCGCCAGUAUCGAGCGCCACAAACGCGGUGCCACCACUGGCCGCCGUCUCCAGCACAACAACCACCUA---------CUCGAUCAGCACAUCCUCGCA ......(((((..((...((((((((((((...(.(((((.(....).))))))...))))).)))))))....)).)))))........---------........((........)). ( -36.20) >consensus AUCCAAUGUUGCAGCACCGGCGGCGCCAGUAUCGAGCGCCACAAACGCGGUGCCACCACUGGCCGCCGUCUCCAGCACAACCGCCACCUACGCCACCAACUCGAUCAGCACAUCCUCGCA .......((((..((...((((((((((((...(.(((((.(....).))))))...))))).)))))))....)).))))..........................((........)). (-32.15 = -32.65 + 0.50)


| Location | 14,974,872 – 14,974,992 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -52.30 |
| Consensus MFE | -46.24 |
| Energy contribution | -48.05 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14974872 120 - 20766785 UGCGAGGAUGUGCUGAUCGAGUUGGUGGCGUAGGUGGCGGUUGUGCUGGAGACGGCGGCCAGUGGUGGCACCGCGUUUGUUGCGCUCGAUACUGGCGCCGCCGGUGCUGCUACAUUGGAU .(((......)))..(((.((((((..(((..((((((.....(((((....)))))((((((((.....))((((.....)))).....))))))))))))..)))..))).))).))) ( -53.10) >DroSim_CAF1 5555 120 - 1 UGCGAGGAUGUGCUGAUCGAGUUGGUGGCGUAGGUGGCGGUUGUGCUGGAGACGGCGGCCAGUGGUGGCACCGCGUUUGUGGCGCUCGAUACUGGCGCCGCCGGUGCUGCAACAUUGGAU .(((......)))...(((((((.(..(((..((((((.....(((((....)))))(((((((..(((.((((....)))).)))...)))))))))))))..)))..)))).)))).. ( -55.90) >DroEre_CAF1 5209 120 - 1 UGCGAGGAUGUGCUAAUCGAGUUGGUGGCGUAGGUGGCGGUUGUGCUAGAGACGGCGGCCAGUGGUGGCACCGCAUUUGUGGCGCUCGAUACUGGCGCCGCCGGUGCUGCAACAUUGGAU .(((......)))...(((((((.(..(((..((((((.(((((.......))))).(((((((..(((.((((....)))).)))...)))))))))))))..)))..)))).)))).. ( -51.10) >DroYak_CAF1 4871 111 - 1 UGCGAGGAUGUGCUGAUCGAG---------UAGGUGGUUGUUGUGCUGGAGACGGCGGCCAGUGGUGGCACCGCGUUUGUGGCGCUCGAUACUGGCGCCGCCGGUGCUGCAACAUUGGAU .((((..((.((((.....))---------)).))..))))..(((((....))))).(((((((..((((((.....((((((((.......))))))))))))))..)..)))))).. ( -49.10) >consensus UGCGAGGAUGUGCUGAUCGAGUUGGUGGCGUAGGUGGCGGUUGUGCUGGAGACGGCGGCCAGUGGUGGCACCGCGUUUGUGGCGCUCGAUACUGGCGCCGCCGGUGCUGCAACAUUGGAU .(((......)))...((((....((((((..((((((.....(((((....)))))(((((((..(((.((((....)))).)))...)))))))))))))..))))))....)))).. (-46.24 = -48.05 + 1.81)

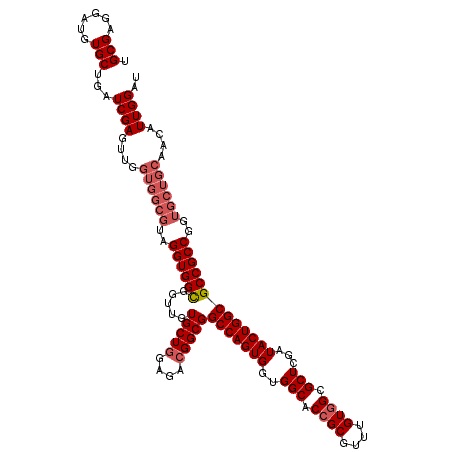
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:09 2006