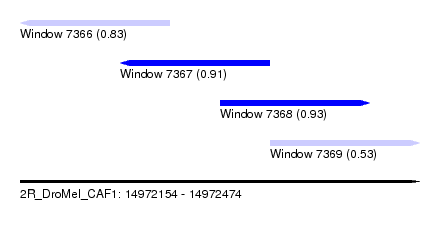
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,972,154 – 14,972,474 |
| Length | 320 |
| Max. P | 0.926257 |

| Location | 14,972,154 – 14,972,274 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -21.79 |
| Energy contribution | -21.60 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.825107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14972154 120 - 20766785 UUAUAAAUAAUUUCCGAAAGGAGCUUGCUGGCUAAAAAUAUACCAUCGAAAUGAAAAUUCCCUUUCGGUGAAAACAACUGUUUUUUAAAAAUUUAAGCACGCACACUAGUUGCAAAUGAA ............(((....)))((.((((..............((((((((.(.......).))))))))(((((....)))))...........)))).)).................. ( -22.10) >DroSim_CAF1 2887 120 - 1 UUAAAAAUAAUUUCCGAAAGGAGCUUGCUGGCUAAAAAUAUACCAUCGAAAUGAAAAUUCCCUUUCGGUGAAAACAACUGUUUUUAAAAAAUUUAAGCACGCACACUAGUGGCAAAUGAA (((((........((((((((((((....))))..........(((....))).......))))))))(((((((....))))))).....)))))((.(((......)))))....... ( -24.40) >DroEre_CAF1 2489 120 - 1 UUAUAAAUAAUUUCCGAAAGGAGCUUGCUGUCUAAAAAUAUACCAUCGAAAUGAAAAUUCCCUUUCGGUGAAAACAACUGUUUUUUAAAAAUUUAAGCACGCACACUAGUGGCAGCAGAA ...........((((....)))).((((((((...........((((((((.(.......).))))))))(((((....)))))..........................)))))))).. ( -26.00) >DroYak_CAF1 2186 120 - 1 UUAUAAAUAAUUUCCGAAAGGAGCUUGCUGGCUAAAAAUAUACCAUUGAAAUGAAAAUUCCCUUUCGGUGAAAACAACUGUUUUUUAAAAAUUUAAGCACGCACACUAGUGGCAGCCGAA .......(((.((((....)))).))).(((((..........((((((((.(.......).))))))))(((((....)))))............((.(((......)))))))))).. ( -23.90) >consensus UUAUAAAUAAUUUCCGAAAGGAGCUUGCUGGCUAAAAAUAUACCAUCGAAAUGAAAAUUCCCUUUCGGUGAAAACAACUGUUUUUUAAAAAUUUAAGCACGCACACUAGUGGCAAAUGAA ............(((....)))((.((((..............((((((((.(.......).))))))))(((((....)))))...........)))).)).................. (-21.79 = -21.60 + -0.19)

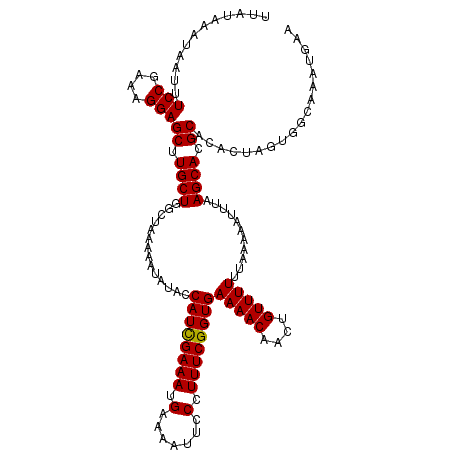
| Location | 14,972,234 – 14,972,354 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -31.12 |
| Energy contribution | -32.38 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14972234 120 - 20766785 AGUGGUCUUUGCACUUACUUAUAUCUGAUUUUGUAAAAGUUUAUCGCGUAAAUCGUUGCGUGGAAACAUUUCGAGGGGGGUUAUAAAUAAUUUCCGAAAGGAGCUUGCUGGCUAAAAAUA ..(((((...(((.(((.(((((.((..(((((.(((.((((.((((((((....)))))))))))).))))))))..)).))))).))).((((....))))..))).)))))...... ( -30.50) >DroSim_CAF1 2967 120 - 1 AGUGGGCUUUGCACAUACUUAUAUCUGCUUUUGUAAAAGUUUAUCGCGUAAAUCGGUGCGUGGAAACAUUUCGAGGGGGGUUAAAAAUAAUUUCCGAAAGGAGCUUGCUGGCUAAAAAUA (((.(((...((.........((.((.((((((.(((.((((.(((((((......))))))))))).))))))))).)).)).........(((....)))))..))).)))....... ( -33.20) >DroEre_CAF1 2569 120 - 1 AGUGGGCUUUGCACAUACUUAUAUCUGCUUUUGUAAAAGUUUAUCGCGUAAAUCGUUGCGUGGAAACAUUUCGAGGGGGGUUAUAAAUAAUUUCCGAAAGGAGCUUGCUGUCUAAAAAUA ((..(((...........(((((.((.((((((.(((.((((.((((((((....)))))))))))).))))))))).)).)))))......(((....))))))..))........... ( -35.00) >DroYak_CAF1 2266 120 - 1 AGUGGGCUUUGCACAUACUUAUAUCUGCUUUUGUAAAAGUUUAUCGCGUAAAUCGUUGCGUGGAAACAUUUCGAGGGGGGUUAUAAAUAAUUUCCGAAAGGAGCUUGCUGGCUAAAAAUA (((.(((...((......(((((.((.((((((.(((.((((.((((((((....)))))))))))).))))))))).)).)))))......(((....)))))..))).)))....... ( -37.00) >consensus AGUGGGCUUUGCACAUACUUAUAUCUGCUUUUGUAAAAGUUUAUCGCGUAAAUCGUUGCGUGGAAACAUUUCGAGGGGGGUUAUAAAUAAUUUCCGAAAGGAGCUUGCUGGCUAAAAAUA (((.(((...((......(((((.((.((((((.(((.((((.((((((((....)))))))))))).))))))))).)).)))))......(((....)))))..))).)))....... (-31.12 = -32.38 + 1.25)

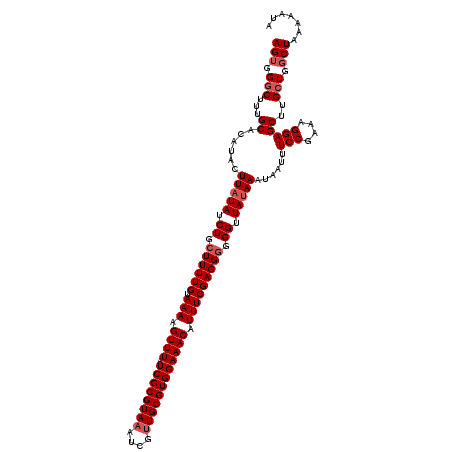
| Location | 14,972,314 – 14,972,434 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -31.01 |
| Energy contribution | -32.08 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.926257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14972314 120 + 20766785 ACUUUUACAAAAUCAGAUAUAAGUAAGUGCAAAGACCACUUGAUUUUCGUAUAGUAUGUAAGUCCACGAAUUGCUUACGUAUUUAUGAAAUAUUACUGUGCGUAGGUCGGUGGGCGUGUU ............................(((....(((((.(((((.(((((((((.((((((.........))))))(((((.....)))))))))))))).))))))))))...))). ( -27.50) >DroSim_CAF1 3047 120 + 1 ACUUUUACAAAAGCAGAUAUAAGUAUGUGCAAAGCCCACUUUAUUUUCGUAUGGUAUGUAAGUCCACGAAUUGCUUACGUAUUUAUGAAAUAUUACUGUGCGUAGGUCGGUGGGCGUGUU .....(((....(((.(((....))).)))...(((((((....(((((((.(((((((((((.........))))))))))))))))))..((((.....))))...)))))))))).. ( -34.20) >DroEre_CAF1 2649 120 + 1 ACUUUUACAAAAGCAGAUAUAAGUAUGUGCAAAGCCCACUUGAUUUUCGUAUGGUAUGUAAGUCCACGAAUUGCUUACGUAUUUAUGAAAUAUUACUGUGCGUAGGUCGGUGGGCGUGUU .....(((....(((.(((....))).)))...(((((((.(((((.(((((((((.((((((.........))))))(((((.....)))))))))))))).))))))))))))))).. ( -37.70) >DroYak_CAF1 2346 120 + 1 ACUUUUACAAAAGCAGAUAUAAGUAUGUGCAAAGCCCACUUGAUUUUCGUAUGCUAUGUAAGUCCACGAAUUGCUUACGUAUUUAUGAAAUAUUACUGUGCGUAGGUCGGUGGGCGUGUA .....((((...(((.(((....))).)))...(((((((.....(((((..(((.....)))..)))))..((((((((((.((........))..)))))))))).))))))).)))) ( -35.00) >consensus ACUUUUACAAAAGCAGAUAUAAGUAUGUGCAAAGCCCACUUGAUUUUCGUAUGGUAUGUAAGUCCACGAAUUGCUUACGUAUUUAUGAAAUAUUACUGUGCGUAGGUCGGUGGGCGUGUU .....(((....(((.(((....))).)))...(((((((.(((((.(((((((((.((((((.........))))))(((((.....)))))))))))))).))))))))))))))).. (-31.01 = -32.08 + 1.06)

| Location | 14,972,354 – 14,972,474 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -31.34 |
| Energy contribution | -31.21 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.98 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14972354 120 + 20766785 UGAUUUUCGUAUAGUAUGUAAGUCCACGAAUUGCUUACGUAUUUAUGAAAUAUUACUGUGCGUAGGUCGGUGGGCGUGUUUCAACGUUUUAGCCCACAGUUUUCACUCAUUUUCCUGCGG (((((((((((.(((((((((((.........)))))))))))))))))).)))).....((((((...((((((................))))))(((....)))......)))))). ( -33.39) >DroSim_CAF1 3087 120 + 1 UUAUUUUCGUAUGGUAUGUAAGUCCACGAAUUGCUUACGUAUUUAUGAAAUAUUACUGUGCGUAGGUCGGUGGGCGUGUUUCAACGUUUUAGCCCACAGUUUUCACUCAUUUUCCUGCGG ....(((((((.(((((((((((.........))))))))))))))))))..........((((((...((((((................))))))(((....)))......)))))). ( -32.79) >DroEre_CAF1 2689 120 + 1 UGAUUUUCGUAUGGUAUGUAAGUCCACGAAUUGCUUACGUAUUUAUGAAAUAUUACUGUGCGUAGGUCGGUGGGCGUGUUUUAUCGUCUCAGCCCACAGUUUUCACUCAUUUUCCUGUGG .(((((.(((((((((.((((((.........))))))(((((.....)))))))))))))).))))).((((((.((...........))))))))......(((..........))). ( -29.90) >DroYak_CAF1 2386 120 + 1 UGAUUUUCGUAUGCUAUGUAAGUCCACGAAUUGCUUACGUAUUUAUGAAAUAUUACUGUGCGUAGGUCGGUGGGCGUGUAUCAGCGUUUCAGCCCACAGUUUUCACUCAUUUUCCUGCGG (((((((((((...(((((((((.........)))))))))..))))))).)))).....((((((...((((((.((...........))))))))(((....)))......)))))). ( -31.40) >consensus UGAUUUUCGUAUGGUAUGUAAGUCCACGAAUUGCUUACGUAUUUAUGAAAUAUUACUGUGCGUAGGUCGGUGGGCGUGUUUCAACGUUUCAGCCCACAGUUUUCACUCAUUUUCCUGCGG ....(((((((.(((((((((((.........))))))))))))))))))..........((((((...((((((................))))))(((....)))......)))))). (-31.34 = -31.21 + -0.13)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:02 2006