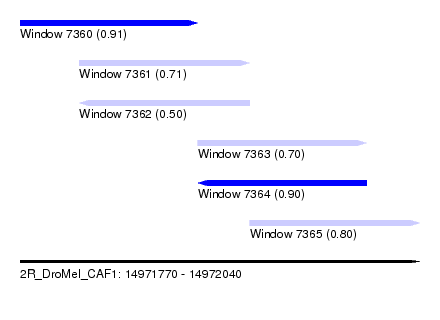
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,971,770 – 14,972,040 |
| Length | 270 |
| Max. P | 0.912913 |

| Location | 14,971,770 – 14,971,890 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.19 |
| Mean single sequence MFE | -36.66 |
| Consensus MFE | -36.42 |
| Energy contribution | -36.48 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14971770 120 + 20766785 UGAGCGAGAGAGCUAAUAGCUUUUCAGCUUUAGCUUUUCUUGGGCCAAUCGGAAAUUGUAUUUCAUUGGUAAGUACAAGAAGUGGCAGCACCAUAACCAGCAAAUCAAUACUUGGGCUGC .((((..((((((.....))))))..))))...(.(((((((.((((((.((((......)))))))))).....))))))).)(((((.(((...................)))))))) ( -34.31) >DroSim_CAF1 1659 120 + 1 UGAGCGAGAGAGCUAAUAGCUUUUCAGCUUUAGCUUUUCUUGGGCCGAUCGGAAAUUGUAUUUGAUUGGUAAGUACAAGAAGUGGCAGCACCAUAACCAGCAAAUCAAUACUUGGGCUGC .((((..((((((.....))))))..))))...(.(((((((.((((((((((.......)))))))))).....))))))).)(((((.(((...................)))))))) ( -37.11) >DroEre_CAF1 2089 120 + 1 UGAGCGAGAGAGCGAAUAGCUUUUCAGCUUUAGCUUUUCUUGGGCCGAUCGGAAAUUAUAUUUGAUUGGUAAGUACAAGAAGUGGCAGCACCAUAACCAGCAAAUCAAUACUUGGGCUGC .((((..((((((.....))))))..))))...(.(((((((.((((((((((.......)))))))))).....))))))).)(((((.(((...................)))))))) ( -37.61) >DroYak_CAF1 1787 120 + 1 UGAGCGAGAGAGCGAAUAGCUUUUCAGCUUUAGCUUUUCUUGGGCCGAUCGGAAAUUGUAUUUGAUUGGUAAGUACAAGAAGUGGCAGCACCAUAACCAGCAAAUCAAUACUUGGGCUGC .((((..((((((.....))))))..))))...(.(((((((.((((((((((.......)))))))))).....))))))).)(((((.(((...................)))))))) ( -37.61) >consensus UGAGCGAGAGAGCGAAUAGCUUUUCAGCUUUAGCUUUUCUUGGGCCGAUCGGAAAUUGUAUUUGAUUGGUAAGUACAAGAAGUGGCAGCACCAUAACCAGCAAAUCAAUACUUGGGCUGC .((((..((((((.....))))))..))))...(.(((((((.((((((((((.......)))))))))).....))))))).)(((((.(((...................)))))))) (-36.42 = -36.48 + 0.06)

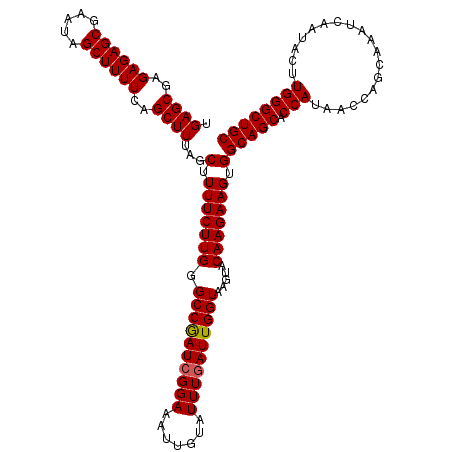
| Location | 14,971,810 – 14,971,925 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.52 |
| Mean single sequence MFE | -34.59 |
| Consensus MFE | -30.55 |
| Energy contribution | -30.86 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14971810 115 + 20766785 UGGGCCAAUCGGAAAUUGUAUUUCAUUGGUAAGUACAAGAAGUGGCAGCACCAUAACCAGCAAAUCAAUACUUGGGCUGCCAUUCCGUUU-----CGUUCCGUUCGUCAGUUUUCCUAGC ..(((.((.(((((.(((((((((....).))))))))(.(((((((((.(((...................)))))))))))).)....-----..))))))).)))............ ( -31.21) >DroSim_CAF1 1699 115 + 1 UGGGCCGAUCGGAAAUUGUAUUUGAUUGGUAAGUACAAGAAGUGGCAGCACCAUAACCAGCAAAUCAAUACUUGGGCUGCCAUUCCGUUU-----CGUUCCGUUCGUUAGUUUUCCUAGC .((..(((.(((((.(((((((((.....)))))))))....(((((((.(((...................)))))))))))))))..)-----))..))....(((((.....))))) ( -37.71) >DroEre_CAF1 2129 120 + 1 UGGGCCGAUCGGAAAUUAUAUUUGAUUGGUAAGUACAAGAAGUGGCAGCACCAUAACCAGCAAAUCAAUACUUGGGCUGCCAUUCCGUUUCGUUUCGUUCCGUUCGUUAGUUUUCCUAGC .((((((((((((.......))))))))))........(.(((((((((.(((...................)))))))))))).).............))....(((((.....))))) ( -32.21) >DroYak_CAF1 1827 120 + 1 UGGGCCGAUCGGAAAUUGUAUUUGAUUGGUAAGUACAAGAAGUGGCAGCACCAUAACCAGCAAAUCAAUACUUGGGCUGCCAUUCCGUUUCGUUUCGUUCCGUUCGUUAGUUUUCCUAGC .((..(((.(((((.(((((((((.....)))))))))(.(((((((((.(((...................)))))))))))).).)))))..)))..))....(((((.....))))) ( -37.21) >consensus UGGGCCGAUCGGAAAUUGUAUUUGAUUGGUAAGUACAAGAAGUGGCAGCACCAUAACCAGCAAAUCAAUACUUGGGCUGCCAUUCCGUUU_____CGUUCCGUUCGUUAGUUUUCCUAGC .((((((((((((.......))))))))))........(.(((((((((.(((...................)))))))))))).).............))....(((((.....))))) (-30.55 = -30.86 + 0.31)

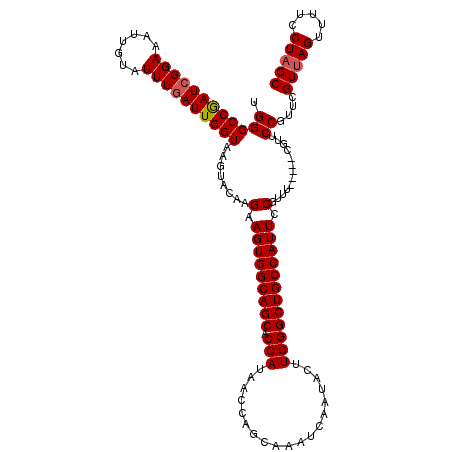
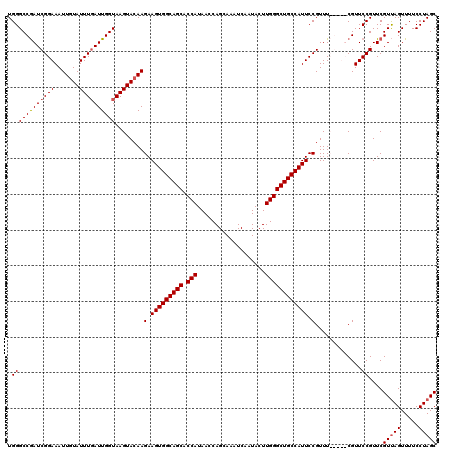
| Location | 14,971,810 – 14,971,925 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.52 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -31.56 |
| Energy contribution | -31.00 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.28 |
| Structure conservation index | 1.01 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14971810 115 - 20766785 GCUAGGAAAACUGACGAACGGAACG-----AAACGGAAUGGCAGCCCAAGUAUUGAUUUGCUGGUUAUGGUGCUGCCACUUCUUGUACUUACCAAUGAAAUACAAUUUCCGAUUGGCCCA (((((.....))).(((.(((((..-----....(((.((((((((((((((......)))).....))).))))))).)))(((((.((.......)).))))).))))).)))))... ( -29.90) >DroSim_CAF1 1699 115 - 1 GCUAGGAAAACUAACGAACGGAACG-----AAACGGAAUGGCAGCCCAAGUAUUGAUUUGCUGGUUAUGGUGCUGCCACUUCUUGUACUUACCAAUCAAAUACAAUUUCCGAUCGGCCCA (((((.....))).(((.(((((..-----....(((.((((((((((((((......)))).....))).))))))).)))(((((.((.......)).))))).))))).)))))... ( -32.00) >DroEre_CAF1 2129 120 - 1 GCUAGGAAAACUAACGAACGGAACGAAACGAAACGGAAUGGCAGCCCAAGUAUUGAUUUGCUGGUUAUGGUGCUGCCACUUCUUGUACUUACCAAUCAAAUAUAAUUUCCGAUCGGCCCA (((((.....))).(((.(((((((........))(((((((((((((((((......)))).....))).))))))).)))........................))))).)))))... ( -30.60) >DroYak_CAF1 1827 120 - 1 GCUAGGAAAACUAACGAACGGAACGAAACGAAACGGAAUGGCAGCCCAAGUAUUGAUUUGCUGGUUAUGGUGCUGCCACUUCUUGUACUUACCAAUCAAAUACAAUUUCCGAUCGGCCCA (((((.....))).(((.(((((...........(((.((((((((((((((......)))).....))).))))))).)))(((((.((.......)).))))).))))).)))))... ( -32.00) >consensus GCUAGGAAAACUAACGAACGGAACG_____AAACGGAAUGGCAGCCCAAGUAUUGAUUUGCUGGUUAUGGUGCUGCCACUUCUUGUACUUACCAAUCAAAUACAAUUUCCGAUCGGCCCA (((((.....))).(((.(((((...........(((.((((((((((((((......)))).....))).))))))).)))(((((.((.......)).))))).))))).)))))... (-31.56 = -31.00 + -0.56)

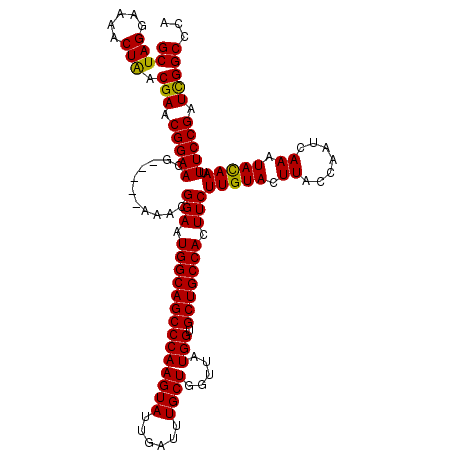
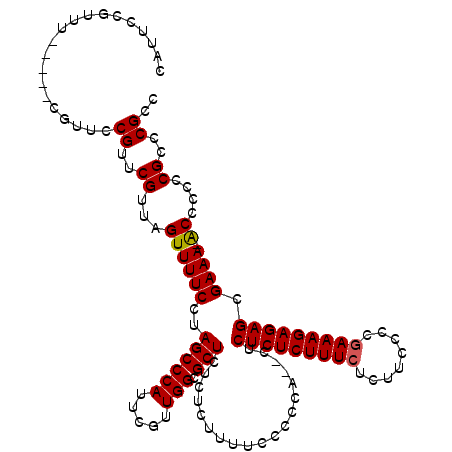
| Location | 14,971,890 – 14,972,004 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.40 |
| Mean single sequence MFE | -22.84 |
| Consensus MFE | -18.72 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14971890 114 + 20766785 CAUUCCGUUU-----CGUUCCGUUCGUCAGUUUUCCUAGCCCAUUCGUUGGCGCUCUCUCUUUUCCGUCAGUCUCUCUCUUUCUCUUCCCCAAAAGAGAGCGAAA-ACCCCCCGCCCGCC ..........-----.....((..((...((((((..((((((.....))).)))(((((((((..(..((.............))..)..))))))))).))))-))....))..)).. ( -20.32) >DroSim_CAF1 1779 112 + 1 CAUUCCGUUU-----CGUUCCGUUCGUUAGUUUUCCUAGCCCAUUCGUUGGCGCUCUCUCUUUUCCCCCA--CUCUCUCUUUCUCUUCCCCGAAAGAGAGCGAAA-ACCCCCCGCCCGCC ..........-----.....((..((...((((((..((((((.....))).)))...............--..(((((((((........))))))))).))))-))....))..)).. ( -23.90) >DroYak_CAF1 1907 120 + 1 CAUUCCGUUUCGUUUCGUUCCGUUCGUUAGUUUUCCUAGCCCAUUCGUUGGCGCUCUUUCUUUUCCCCCACACUCUCUCUUUCUCUCUCCCGAAAGAGAGCGAAAAGCCCCCCGCCCGCC .........................(((((.....)))))......((.((((......((((((.........(((((((((........))))))))).)))))).....)))).)). ( -24.30) >consensus CAUUCCGUUU_____CGUUCCGUUCGUUAGUUUUCCUAGCCCAUUCGUUGGCGCUCUCUCUUUUCCCCCA__CUCUCUCUUUCUCUUCCCCGAAAGAGAGCGAAA_ACCCCCCGCCCGCC ....................((..((...((((((..((((((.....))).)))...................(((((((((........))))))))).)))).))....))..)).. (-18.72 = -18.83 + 0.11)

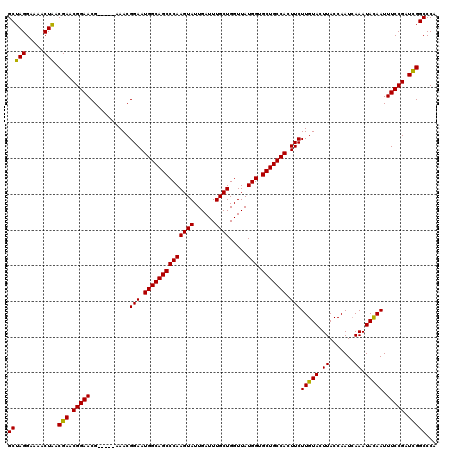
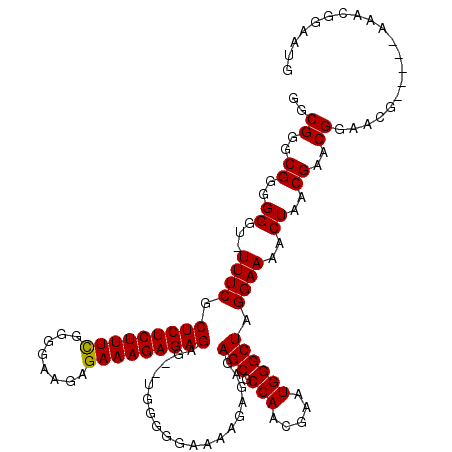
| Location | 14,971,890 – 14,972,004 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.40 |
| Mean single sequence MFE | -31.41 |
| Consensus MFE | -28.92 |
| Energy contribution | -28.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14971890 114 - 20766785 GGCGGGCGGGGGGU-UUUCGCUCUCUUUUGGGGAAGAGAAAGAGAGAGACUGACGGAAAAGAGAGAGCGCCAACGAAUGGGCUAGGAAAACUGACGAACGGAACG-----AAACGGAAUG ..((..((...(((-((((((((((((((..(..((.............))..)....))))))))))(((........)))...)))))))..))..)).....-----.......... ( -30.82) >DroSim_CAF1 1779 112 - 1 GGCGGGCGGGGGGU-UUUCGCUCUCUUUCGGGGAAGAGAAAGAGAGAG--UGGGGGAAAAGAGAGAGCGCCAACGAAUGGGCUAGGAAAACUAACGAACGGAACG-----AAACGGAAUG ..((..((...(((-((((.(((((((((........)))))))))..--...............(((.(((.....))))))..)))))))..))..)).....-----.......... ( -32.80) >DroYak_CAF1 1907 120 - 1 GGCGGGCGGGGGGCUUUUCGCUCUCUUUCGGGAGAGAGAAAGAGAGAGUGUGGGGGAAAAGAAAGAGCGCCAACGAAUGGGCUAGGAAAACUAACGAACGGAACGAAACGAAACGGAAUG ..((..((..((..(((((.(((((((((........)))))))))...................(((.(((.....)))))).))))).))..))..)).................... ( -30.60) >consensus GGCGGGCGGGGGGU_UUUCGCUCUCUUUCGGGGAAGAGAAAGAGAGAG__UGGGGGAAAAGAGAGAGCGCCAACGAAUGGGCUAGGAAAACUAACGAACGGAACG_____AAACGGAAUG ..((..((..((...((((.(((((((((........)))))))))...................(((.(((.....)))))).))))..))..))..)).................... (-28.92 = -28.70 + -0.22)


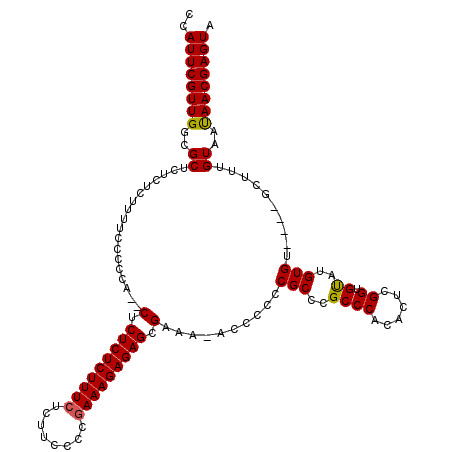
| Location | 14,971,925 – 14,972,040 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -25.37 |
| Energy contribution | -25.27 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14971925 115 + 20766785 CCAUUCGUUGGCGCUCUCUCUUUUCCGUCAGUCUCUCUCUUUCUCUUCCCCAAAAGAGAGCGAAA-ACCCCCCGCCCGCCCACACUCGGUGUAUGUGU----GCUUUGUAAUAACGAGUA ..((((((((..((............((...((.((((((((..........)))))))).))..-)).....((.(((..((((...))))..))).----))...))..)))))))). ( -25.70) >DroSim_CAF1 1814 113 + 1 CCAUUCGUUGGCGCUCUCUCUUUUCCCCCA--CUCUCUCUUUCUCUUCCCCGAAAGAGAGCGAAA-ACCCCCCGCCCGCCCACACUCGGUGUAUGUGU----GCUUUGUAACAACGAGUA ..((((((((..((................--(.(((((((((........))))))))).)...-.......((.(((..((((...))))..))).----))...))..)))))))). ( -31.20) >DroYak_CAF1 1947 120 + 1 CCAUUCGUUGGCGCUCUUUCUUUUCCCCCACACUCUCUCUUUCUCUCUCCCGAAAGAGAGCGAAAAGCCCCCCGCCCGCCCCCACUUGGUGCAUGUGUGCGUGCUUUGUAAUAACGAGUA ..(((((((((((......((((((.........(((((((((........))))))))).)))))).....)))).(((.......)))(((((....)))))........))))))). ( -32.70) >consensus CCAUUCGUUGGCGCUCUCUCUUUUCCCCCA__CUCUCUCUUUCUCUUCCCCGAAAGAGAGCGAAA_ACCCCCCGCCCGCCCACACUCGGUGUAUGUGU____GCUUUGUAAUAACGAGUA ..((((((((..((..................(.(((((((((........))))))))).)..........(((..((((......)).))..)))..........))..)))))))). (-25.37 = -25.27 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:58 2006