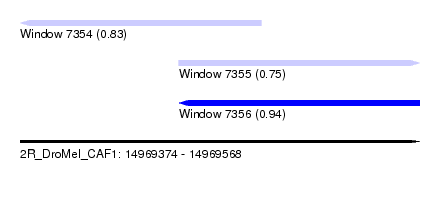
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,969,374 – 14,969,568 |
| Length | 194 |
| Max. P | 0.937703 |

| Location | 14,969,374 – 14,969,491 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.96 |
| Mean single sequence MFE | -44.52 |
| Consensus MFE | -36.36 |
| Energy contribution | -35.56 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14969374 117 - 20766785 UCGCCGGCUGGACACGCGGGGCAACCACUGUAGAGGCAUCUCCAUAGUUCGAGGACCAAGC---CUGCUGUUCCAUCCGUUGCUUCGACAGAUGCAACGAGUCCAGGGACAGCAAAGAAG .....((((((.(...((((....))(((((.(((....))).))))).))..).)).)))---)(((((((((..(((((((.((....)).)))))).)....)))))))))...... ( -44.60) >DroSec_CAF1 8955 117 - 1 UCGCCGGCUGGACUCGCGGUGCAACCACUGUAGAGGCAUCACCAUAGUUCGAAGACCAAGC---CUGCUGUUCCAACCGUUGCUUCGACAGAUGCAACGAGUCCAGGGACAGCACAGAAG .....((((((.(((((((((....)))))).)))...((..........))...)).)))---)(((((((((.((((((((.((....)).)))))).))...)))))))))...... ( -41.50) >DroSim_CAF1 9688 117 - 1 UCGCCGGCUGGACUCGCGGUGCAACCACUGUAGAGGCAUCACCAUAGUUCGAAGACCAAGC---CUGCUGUUCCAUCCGUUGCUUCGACAGAUGCAACGAGUCCAGGGACAGCAGAGAAG .....((((((.(((((((((....)))))).)))...((..........))...)).)))---)(((((((((..(((((((.((....)).)))))).)....)))))))))...... ( -40.70) >DroEre_CAF1 9095 117 - 1 UCGCUGGCUGGACUCGCGGAGCAACCACAGUAGAGGCACCUGCAUAGUCCGAGGACCAAGC---CUGCUGUUCCCUCCGUUGCUUUGACAGAUGCAACGAGUCCAGGGACAGCAGAGAGG ..((((.((((((((..((((.....(((((((..((....))...(((....))).....---)))))))...))))(((((..........)))))))))))))...))))....... ( -46.90) >DroYak_CAF1 9382 120 - 1 UCGCUGGCUGGGCUCGCGGGGCAAUCACUGUAGAGGCACCUCCAUAGUUUGAAGACCAAGCCGCCUGCUGUUCCAUCCGUUGCUUCGACAGAUGCAACGAGUCCAGGGACAGCAGAGAAG ..((.(((((((((.....)))....(((((.(((....))).))))).......)).))))))((((((((((..(((((((.((....)).)))))).)....))))))))))..... ( -48.90) >consensus UCGCCGGCUGGACUCGCGGGGCAACCACUGUAGAGGCAUCUCCAUAGUUCGAAGACCAAGC___CUGCUGUUCCAUCCGUUGCUUCGACAGAUGCAACGAGUCCAGGGACAGCAGAGAAG .....(((((((((...((((...((........))...))))..)))))..)).)).......((((((((((..(((((((.((....)).)))))).)....))))))))))..... (-36.36 = -35.56 + -0.80)


| Location | 14,969,451 – 14,969,568 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -39.45 |
| Consensus MFE | -31.26 |
| Energy contribution | -31.14 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14969451 117 + 20766785 GAUGCCUCUACAGUGGUUGCCCCGCGUGUCCAGCCGGCGAUGAUGUUUCAACAGAAACCCAUGAGA---CCAUUAGAGUCCCAAUGGUAUCGCCAUGGACAAAGUAACCUGGCCCAAGGC ...((((.......((((((......((((((...(((((((..(((((....)))))((((..((---((....).)))...))))))))))).))))))..)))))).......)))) ( -39.84) >DroSec_CAF1 9032 120 + 1 GAUGCCUCUACAGUGGUUGCACCGCGAGUCCAGCCGGCGAUGACGUUUCAACAGAAACACAUGAGAAACCCAUUAGAGUCCCAAUGGUAUCGCCAUGGACAAAGUAACCUGGCACAAGGC ..((((..(((.((((.....))))..(((((...(((((((..(((((....)))))...........(((((.(....).)))))))))))).)))))...)))....))))...... ( -38.80) >DroSim_CAF1 9765 120 + 1 GAUGCCUCUACAGUGGUUGCACCGCGAGUCCAGCCGGCGAUGAUGUUUCAACAGAAACACAUGAGAAACCCAUUGGAGUCGCAAUGGCAUCGCCAUGGACAAAGUAACCUGGCACAAGGC ..((((..(((.((((.....))))..(((((...(((((((.((((((....))))))..........((((((......))))))))))))).)))))...)))....))))...... ( -44.20) >DroEre_CAF1 9172 117 + 1 GGUGCCUCUACUGUGGUUGCUCCGCGAGUCCAGCCAGCGAUGGUGUUUCAACAGAAACCCAUGAGG---CCAUUAGAGUCCCAAUGGUAUCGUAAUGGACAAAAUAACCUAGCCCAAAGC ((.((......(((((.....))))).(((((....(((((((.(((((....))))))).....(---(((((.(....).)))))))))))..)))))...........))))..... ( -34.10) >DroYak_CAF1 9462 117 + 1 GGUGCCUCUACAGUGAUUGCCCCGCGAGCCCAGCCAGCGAUGAUGUUCCAACAGAAGCCCAUGAGG---CCAUUAGAGUCACAAUGGUAUCGCCAUGGGCCAAGUAAUCUGGCCCAAGGC ((((((......((((((....(((..((...))..))).((((((((.....)))(((.....))---)))))).))))))...))))))(((.(((((((.......))))))).))) ( -40.30) >consensus GAUGCCUCUACAGUGGUUGCACCGCGAGUCCAGCCGGCGAUGAUGUUUCAACAGAAACCCAUGAGA___CCAUUAGAGUCCCAAUGGUAUCGCCAUGGACAAAGUAACCUGGCCCAAGGC ...((((.......((((((.......(((((...(((((((..(((((....)))))...........(((((........)))))))))))).)))))...)))))).......)))) (-31.26 = -31.14 + -0.12)

| Location | 14,969,451 – 14,969,568 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -44.36 |
| Consensus MFE | -36.16 |
| Energy contribution | -35.92 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14969451 117 - 20766785 GCCUUGGGCCAGGUUACUUUGUCCAUGGCGAUACCAUUGGGACUCUAAUGG---UCUCAUGGGUUUCUGUUGAAACAUCAUCGCCGGCUGGACACGCGGGGCAACCACUGUAGAGGCAUC (((((....(((.......(((((((((((((.((((.((((((.....))---))))))))(((((....)))))...)))))))..))))))....((....)).)))..)))))... ( -49.10) >DroSec_CAF1 9032 120 - 1 GCCUUGUGCCAGGUUACUUUGUCCAUGGCGAUACCAUUGGGACUCUAAUGGGUUUCUCAUGUGUUUCUGUUGAAACGUCAUCGCCGGCUGGACUCGCGGUGCAACCACUGUAGAGGCAUC (((((...............((((((((((((.(((((((....))))))).........(((((((....)))))).))))))))..)))))..((((((....)))))).)))))... ( -43.40) >DroSim_CAF1 9765 120 - 1 GCCUUGUGCCAGGUUACUUUGUCCAUGGCGAUGCCAUUGCGACUCCAAUGGGUUUCUCAUGUGUUUCUGUUGAAACAUCAUCGCCGGCUGGACUCGCGGUGCAACCACUGUAGAGGCAUC (((((...............(((((((((((((((((((......)))))).........(((((((....)))))))))))))))..)))))..((((((....)))))).)))))... ( -45.40) >DroEre_CAF1 9172 117 - 1 GCUUUGGGCUAGGUUAUUUUGUCCAUUACGAUACCAUUGGGACUCUAAUGG---CCUCAUGGGUUUCUGUUGAAACACCAUCGCUGGCUGGACUCGCGGAGCAACCACAGUAGAGGCACC (((((..(((.((((.....(((((.........((((((....))))))(---((..(((((((((....))))).))))....))))))))..((...))))))..))).)))))... ( -35.70) >DroYak_CAF1 9462 117 - 1 GCCUUGGGCCAGAUUACUUGGCCCAUGGCGAUACCAUUGUGACUCUAAUGG---CCUCAUGGGCUUCUGUUGGAACAUCAUCGCUGGCUGGGCUCGCGGGGCAAUCACUGUAGAGGCACC (((.((((((((.....)))))))).))).........(((.(((((...(---((((.(((((((..((..(..........)..)).))))))).)))))........))))).))). ( -48.20) >consensus GCCUUGGGCCAGGUUACUUUGUCCAUGGCGAUACCAUUGGGACUCUAAUGG___UCUCAUGGGUUUCUGUUGAAACAUCAUCGCCGGCUGGACUCGCGGGGCAACCACUGUAGAGGCAUC (((((......((((.....((((((((((((.(((((((....)))))))...........(((((....)))))...)))))))..)))))..((...))))))......)))))... (-36.16 = -35.92 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:50 2006