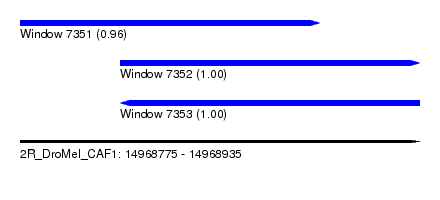
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,968,775 – 14,968,935 |
| Length | 160 |
| Max. P | 0.999983 |

| Location | 14,968,775 – 14,968,895 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -37.06 |
| Consensus MFE | -33.92 |
| Energy contribution | -33.16 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14968775 120 + 20766785 CAUUUUCCUGGAACAGCGCGAAAUGAUAGCCAAACUUUUAUUGUAUGGCCGUACCUUUGGCAAGCUCUUGCUGUAUUCGCUUGGCAUAGGCCAACAGCAAGUGCAGCAAGCAACCUUCGC .........((...(((((.(((.((..((((.((.......)).))))..).).))).))..)))(((((((((((.(((((((....))))..))).)))))))))))...))..... ( -37.60) >DroSec_CAF1 8356 120 + 1 CAUUUUCCUGGAACAGCGCGAACUGACAAGCAAACUUUUAUUGUACGGCCGUACCUUUGGCAAACUCUUGCUGUAUUCGCUGGGCUUAAGCCAACAGCAGGUGCAGCAAGCAACCUUCGC .................(((((.......((((.......))))...((((......)))).....((((((((((..(((((((....)))..))))..)))))))))).....))))) ( -36.50) >DroSim_CAF1 9089 120 + 1 CAUUUUCCUGGAACAGCGCGAACUGACAAGCAAACUUUUAUUGUACGGCCGUACCUUUGGCAAACUCUUGCUGUAUUCGCUGGGCUUAAGCCAACAGCAGGUGCAGCAAGCAACCUUCGC .................(((((.......((((.......))))...((((......)))).....((((((((((..(((((((....)))..))))..)))))))))).....))))) ( -36.50) >DroEre_CAF1 8498 120 + 1 CGUUUUCCUGGAACAGCGUGAAAUGACAGCAAAACUUUUAUUGUACAGCCGCACCUUUGGCAAGCUCUUGCUGUAUUCGCUGGGUUUAAGCCAACAGCAAGUGCAGCAGGCCACCUUCGC ........(((...((((((.(((((.((.....)).))))).))).((((......))))..)))(((((((((((.(((((((....)))..)))).))))))))))))))....... ( -36.30) >DroYak_CAF1 8782 120 + 1 ACUUUUCCUGGAGCAGCGUGAAAUGACAGCCAAACUUUUAUUGUACGGCCGCACCUUUGGCAAGCUCUUGCUGUAUUCGCUGGGUUUGAGCCAACAGCAAGUGCAGCAGGCAACCUUCGC ......((((((((..((((.(((((.((.....)).))))).))))((((......))))..))))).((((((((.(((((((....)))..)))).))))))))))).......... ( -38.40) >consensus CAUUUUCCUGGAACAGCGCGAAAUGACAGCCAAACUUUUAUUGUACGGCCGUACCUUUGGCAAGCUCUUGCUGUAUUCGCUGGGCUUAAGCCAACAGCAAGUGCAGCAAGCAACCUUCGC .................(((((.........................((((......)))).....(((((((((((.(((((((....)))..)))).))))))))))).....))))) (-33.92 = -33.16 + -0.76)


| Location | 14,968,815 – 14,968,935 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.58 |
| Mean single sequence MFE | -43.82 |
| Consensus MFE | -41.76 |
| Energy contribution | -41.28 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.95 |
| SVM decision value | 5.32 |
| SVM RNA-class probability | 0.999983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14968815 120 + 20766785 UUGUAUGGCCGUACCUUUGGCAAGCUCUUGCUGUAUUCGCUUGGCAUAGGCCAACAGCAAGUGCAGCAAGCAACCUUCGCAUCCUGUUUUAUUGGGAUCUACCCAUAUGCCAUGCUGGCC ......(((((((....(((((.(((..(((((((((.(((((((....))))..))).)))))))))))).....................((((.....))))..)))))))).)))) ( -42.90) >DroSec_CAF1 8396 120 + 1 UUGUACGGCCGUACCUUUGGCAAACUCUUGCUGUAUUCGCUGGGCUUAAGCCAACAGCAGGUGCAGCAAGCAACCUUCGCAUCGUGUUUUAUUGGGAUCUAUCCAUAUGCCAUGCUGGCC ......(((((((....(((((....((((((((((..(((((((....)))..))))..))))))))))........................(((....)))...)))))))).)))) ( -42.90) >DroSim_CAF1 9129 120 + 1 UUGUACGGCCGUACCUUUGGCAAACUCUUGCUGUAUUCGCUGGGCUUAAGCCAACAGCAGGUGCAGCAAGCAACCUUCGCAUCCUGCUUUAUUGGGAUCUACCCAUAUGCCAUGCUGGCC ......(((((((....(((((....((((((((((..(((((((....)))..))))..))))))))))........((.....)).....((((.....))))..)))))))).)))) ( -46.00) >DroEre_CAF1 8538 120 + 1 UUGUACAGCCGCACCUUUGGCAAGCUCUUGCUGUAUUCGCUGGGUUUAAGCCAACAGCAAGUGCAGCAGGCCACCUUCGCAUCCUGCUUUAUUGGCGUCUAUCCAUAUGCCAUGCUGGCC .......((((((....(((((.....((((((((((.(((((((....)))..)))).))))))))))((((.....((.....)).....))))...........)))))))).))). ( -41.70) >DroYak_CAF1 8822 120 + 1 UUGUACGGCCGCACCUUUGGCAAGCUCUUGCUGUAUUCGCUGGGUUUGAGCCAACAGCAAGUGCAGCAGGCAACCUUCGCGUCCUGCUUUAUUGGGCUCUACCCAUAUGCCAUGCUGGCC ......(((((((....((((((((....)))........(((((..(((((....((....))(((((((.........).))))))......))))).)))))..)))))))).)))) ( -45.60) >consensus UUGUACGGCCGUACCUUUGGCAAGCUCUUGCUGUAUUCGCUGGGCUUAAGCCAACAGCAAGUGCAGCAAGCAACCUUCGCAUCCUGCUUUAUUGGGAUCUACCCAUAUGCCAUGCUGGCC ......(((((((....(((((....(((((((((((.(((((((....)))..)))).)))))))))))......................((((.....))))..)))))))).)))) (-41.76 = -41.28 + -0.48)

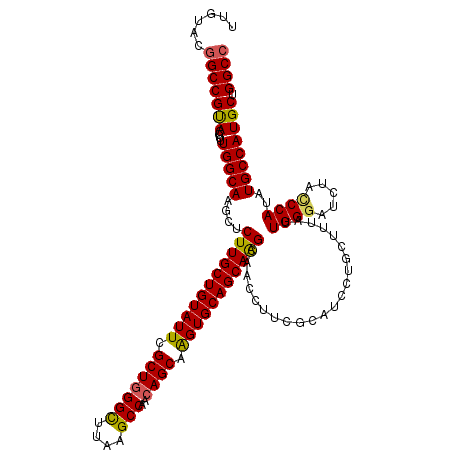
| Location | 14,968,815 – 14,968,935 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.58 |
| Mean single sequence MFE | -44.72 |
| Consensus MFE | -39.60 |
| Energy contribution | -40.64 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.07 |
| SVM RNA-class probability | 0.999785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14968815 120 - 20766785 GGCCAGCAUGGCAUAUGGGUAGAUCCCAAUAAAACAGGAUGCGAAGGUUGCUUGCUGCACUUGCUGUUGGCCUAUGCCAAGCGAAUACAGCAAGAGCUUGCCAAAGGUACGGCCAUACAA ((((.((.((((...((((.....))))................(((((.(((((((...(((((..((((....)))))))))...))))))))))))))))...))..))))...... ( -43.30) >DroSec_CAF1 8396 120 - 1 GGCCAGCAUGGCAUAUGGAUAGAUCCCAAUAAAACACGAUGCGAAGGUUGCUUGCUGCACCUGCUGUUGGCUUAAGCCCAGCGAAUACAGCAAGAGUUUGCCAAAGGUACGGCCGUACAA ((((.((.((((...(((.......)))...((((.....((....))..(((((((....(((((..(((....))))))))....))))))).))))))))...))..))))...... ( -39.00) >DroSim_CAF1 9129 120 - 1 GGCCAGCAUGGCAUAUGGGUAGAUCCCAAUAAAGCAGGAUGCGAAGGUUGCUUGCUGCACCUGCUGUUGGCUUAAGCCCAGCGAAUACAGCAAGAGUUUGCCAAAGGUACGGCCGUACAA ((((.((.(((((..(((((..(..(((((..((((((.(((((((....)))..)))))))))))))))..)..)))))((.......)).......)))))...))..))))...... ( -43.60) >DroEre_CAF1 8538 120 - 1 GGCCAGCAUGGCAUAUGGAUAGACGCCAAUAAAGCAGGAUGCGAAGGUGGCCUGCUGCACUUGCUGUUGGCUUAAACCCAGCGAAUACAGCAAGAGCUUGCCAAAGGUGCGGCUGUACAA ((((.((((((((..(((.......)))....((((((..((....))..))))))((.(((((((((.(((.......))).)..)))))))).)).)))))....)))))))...... ( -45.60) >DroYak_CAF1 8822 120 - 1 GGCCAGCAUGGCAUAUGGGUAGAGCCCAAUAAAGCAGGACGCGAAGGUUGCCUGCUGCACUUGCUGUUGGCUCAAACCCAGCGAAUACAGCAAGAGCUUGCCAAAGGUGCGGCCGUACAA ((((.((((((((..((((.....))))....((((((..((....))..))))))((.(((((((((.(((.......))).)..)))))))).)).)))))....)))))))...... ( -52.10) >consensus GGCCAGCAUGGCAUAUGGGUAGAUCCCAAUAAAGCAGGAUGCGAAGGUUGCUUGCUGCACUUGCUGUUGGCUUAAGCCCAGCGAAUACAGCAAGAGCUUGCCAAAGGUACGGCCGUACAA ((((.((.(((((..((((.....))))....((((((..((....))..))))))((.(((((((((.(((.......))).)..)))))))).)).)))))...))..))))...... (-39.60 = -40.64 + 1.04)

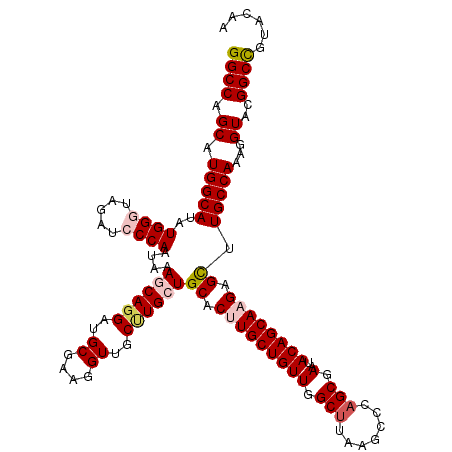
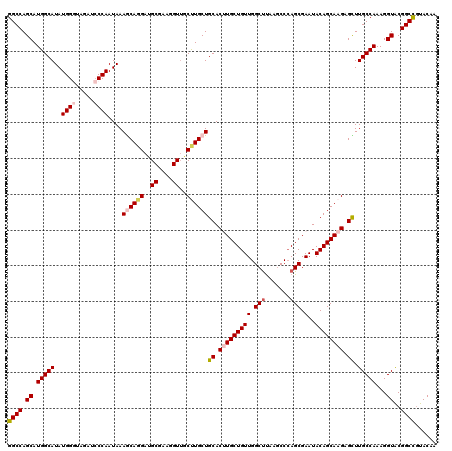
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:47 2006