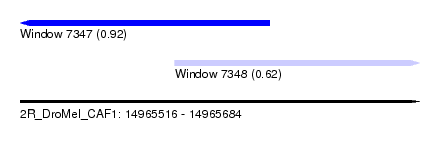
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,965,516 – 14,965,684 |
| Length | 168 |
| Max. P | 0.924225 |

| Location | 14,965,516 – 14,965,621 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.95 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -11.70 |
| Energy contribution | -12.20 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
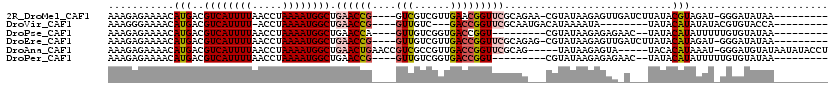
>2R_DroMel_CAF1 14965516 105 - 20766785 AAAGAGAAAACAUGACGUCAUUUUAACCUAAAAUGGCUGAACCG----GUCGUCGUUGAACGGUUCGCAGAA-CGUAUAAGAGUUGAUCUUAUACGUAGAU-GGGAUAUAA--------- ..........(((...(((((((((...))))))))).((((((----.(((....))).)))))).....(-(((((((((.....))))))))))..))-)........--------- ( -30.10) >DroVir_CAF1 2610 95 - 1 AAAGGGAAAACAUGACGUCAUUUU-ACCUAAAAUGGCUGAACCG----GUUGUC---GACCGGUUCGCAAUGACAUAAAAUA--------UAUACAUAUAUACGUGUACCA--------- ...((....(((((..((((((..-.((......))..((((((----(((...---)))))))))..)))))).....(((--------((....))))).))))).)).--------- ( -26.70) >DroPse_CAF1 5661 96 - 1 AAAGAGAAAACAUGACGUCAUUUUAACCUAAAAUGGCUGAACCA----GUUGUCGGUGACCGGU---------CGUAUAAGAGAGAAC--UAUACAUAUUUUUGUGUAUAA--------- ...........((((((((((((((...)))))))))...(((.----......))).....))---------)))............--((((((((....)))))))).--------- ( -19.40) >DroEre_CAF1 5480 105 - 1 AAAGAGAAAACAUGACGUCAUUUUAACCUAAAAUGGCUGAACCG----GUUGUCGUUGACCGGUUCGCAGAG-CGUAUAAGAGUUGAUCUUAUACAUAGAU-GGGAUAUAA--------- ..........(((...(((((((((...))))))))).((((((----(((......)))))))))......-.((((((((.....))))))))....))-)........--------- ( -30.90) >DroAna_CAF1 4909 109 - 1 AAAGAGAAAACAUGACGUCAUUUUAACCUAAAAUGGCUGAACUGAACCGUCGCCGUUGACCGGUUCGCAG-----UAUAAGAGUA-----UACACAUAAAU-GGGAUGUAUAAUAUACCU .........((((..((((((((((...)))))))))......(((((((((....))).))))))...(-----((((....))-----)))........-)..))))........... ( -21.80) >DroPer_CAF1 5584 96 - 1 AAAGAGAAAACAUGACGUCAUUUUAACCUAAAAUGGCUGAACCG----GUUGUCGGUGACCGGU---------CGUAUAAGAGAGAAC--UAUACAUAUUUUUGUGUAUAA--------- ...........((((((((((((((...))))))))).....((----(((......)))))))---------)))............--((((((((....)))))))).--------- ( -23.90) >consensus AAAGAGAAAACAUGACGUCAUUUUAACCUAAAAUGGCUGAACCG____GUUGUCGUUGACCGGUUCGCAG___CGUAUAAGAGUG_A___UAUACAUAUAU_GGGAUAUAA_________ ...........(((..((((((((.....)))))))).((((((....(((......)))))))))............................)))....................... (-11.70 = -12.20 + 0.50)
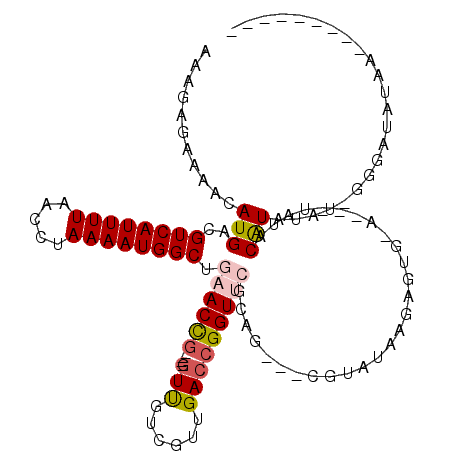
| Location | 14,965,581 – 14,965,684 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.28 |
| Mean single sequence MFE | -18.90 |
| Consensus MFE | -17.29 |
| Energy contribution | -17.49 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14965581 103 + 20766785 UCAGCCAUUUUAGGUUAAAAUGACGUCAUGUUUUCUCUUUAGAAGACGCAUAGCGGCAAAAAAAAAAAAAUUAAAUGA------AAAAACCGGCCGACGCUCACACAUA ..((((......))))....(((((((..(((((((....))))))).......(((.....................------........))))))).)))...... ( -18.65) >DroSec_CAF1 5446 102 + 1 UCAGCCAUUUUAGGUUAAAAUGACGUCAUGUUUUCUCUUUAGAAGACGCAUAGCGGCAAAAAA-AAAAAAUAAAAUGA------AAAAACCGGCCGACGCUCACACAUA ..((((......))))....(((((((..(((((((....))))))).......(((......-..............------........))))))).)))...... ( -18.69) >DroSim_CAF1 6161 99 + 1 UCAGCCAUUUUAGGUUAAAAUGACGUCAUGUUUUCUCUUUAGAAGACGCAUAGCGGCAAAAA----AAAAUAAAAUGA------AAAAACCGGCCGACGCUCACACAUA ..((((......))))....(((((((..(((((((....))))))).......(((.....----............------........))))))).)))...... ( -18.81) >DroEre_CAF1 5545 90 + 1 UCAGCCAUUUUAGGUUAAAAUGACGUCAUGUUUUCUCUUUAGAAGACGCAUAGCGGAAA-------------------AAAAGAAUAAACCGGCCGACGCUCACACAUA ..((((......))))....(((((((..(((((((....)))))))......(((...-------------------...........)))...)))).)))...... ( -17.64) >DroAna_CAF1 4978 82 + 1 UCAGCCAUUUUAGGUUAAAAUGACGUCAUGUUUUCUCUUUAGAAGACGCAUAGCGGCAA---------------------------AAACCGGCCGACGCUCACCAAUA ..((((......))))....(((((((..(((((((....))))))).......(((..---------------------------......))))))).)))...... ( -20.70) >consensus UCAGCCAUUUUAGGUUAAAAUGACGUCAUGUUUUCUCUUUAGAAGACGCAUAGCGGCAAAAA____AAAAU_AAAUGA______AAAAACCGGCCGACGCUCACACAUA ..((((......))))....(((((((..(((((((....))))))).......(((...................................))))))).)))...... (-17.29 = -17.49 + 0.20)

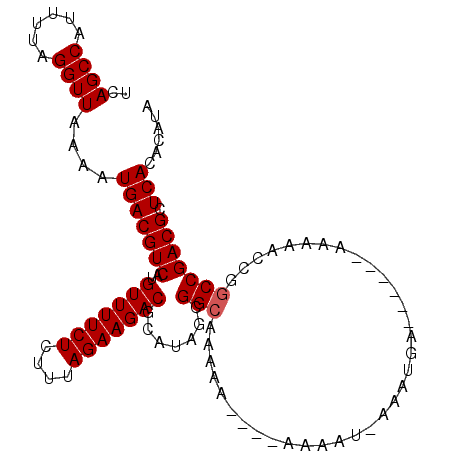
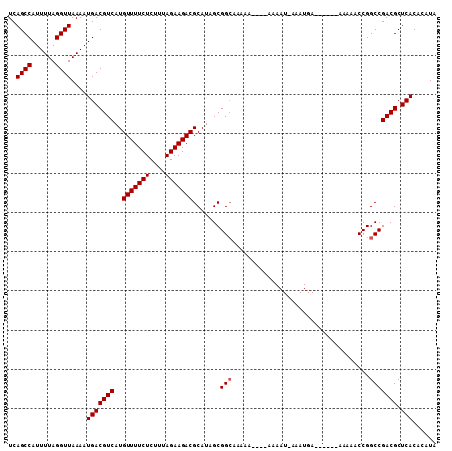
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:42 2006