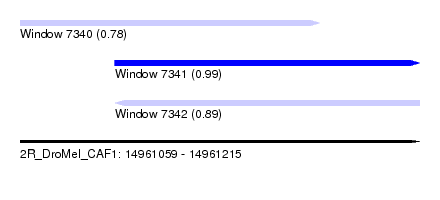
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,961,059 – 14,961,215 |
| Length | 156 |
| Max. P | 0.993343 |

| Location | 14,961,059 – 14,961,176 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 94.18 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -17.04 |
| Energy contribution | -16.24 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14961059 117 + 20766785 AAUAUUUAUAUUUUAUCUUACUUGUCAAAUAUCAGUCAGUGACACCCACACACCGCCGCACGGUACGCACACACACACACAAUCAAUACACACUGUGCACACUGUUUUUGGCGUCCC .......................((((((...((((..(((.....)))..((((.....))))..(((((......................)))))..))))..))))))..... ( -19.95) >DroSec_CAF1 976 117 + 1 AAUAUUUAUAUUUUAUCUUACUUGUCAAAUAUCAGUCAGUGACACCCACACACGGCCGCACGGUACACACACACACACGCAAUCAAUACACACUGUGCACACUGUUUUUGGCGGCCC .....................((((((((...((((..(((.....)))........(((((((...........................)))))))..))))..))))))))... ( -20.23) >DroSim_CAF1 1173 117 + 1 AAUAUUUAUAUUUUAUCUUACUUGUCAAAUAUCAGUCAGUGACACCCACACACGGCCGCACGGUACACACACACACACGCAAUCAAUACACACUGUGCACACUGUUUUCGGCGGCCC ......................(((((............))))).........((((((.(((.(((....((((..................)))).....)))..))))))))). ( -20.37) >DroEre_CAF1 1054 115 + 1 AUUAUUUAUAUUUUAUCUUACUUGUCAAAUAUCAGUCAGUGACACUCACACACGGCCGCACGAAACGCACACA--CACACAAUCAAUACACACUGUGCACACUGUUUUCGGUGGCCC ......................................(((.....)))....((((((.(((((.(((....--..((((............)))).....)))))))))))))). ( -21.60) >DroYak_CAF1 1235 115 + 1 AUUAUUUAUAUUUUAUCUUACUUGUCAAAUAUCAGUCAGUGACACUCACACACGGCCGCACGAAACGCACACA--CACACAAUCAAUACACACUGUGCACACUGUUUUCGGUGGCCC ......................................(((.....)))....((((((.(((((.(((....--..((((............)))).....)))))))))))))). ( -21.60) >consensus AAUAUUUAUAUUUUAUCUUACUUGUCAAAUAUCAGUCAGUGACACCCACACACGGCCGCACGGUACGCACACACACACACAAUCAAUACACACUGUGCACACUGUUUUCGGCGGCCC ......................(((((............))))).........((((((.(((...(((......((((..............)))).....)))..))))))))). (-17.04 = -16.24 + -0.80)

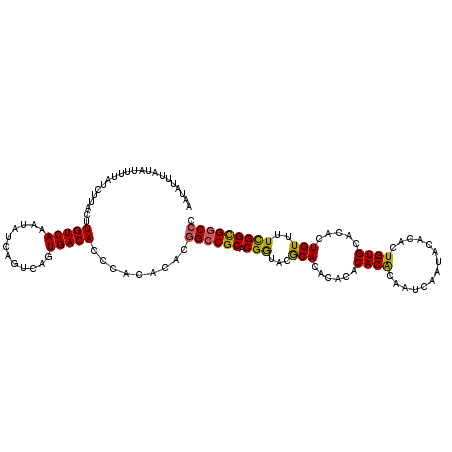
| Location | 14,961,096 – 14,961,215 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 92.09 |
| Mean single sequence MFE | -33.22 |
| Consensus MFE | -29.14 |
| Energy contribution | -27.82 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14961096 119 + 20766785 AGUGACACCCACACACCGCCGCACGGUACGCACACACACACACAAUCAAUACACACUGUGCACACUGUUUUUGGCGUCCCUGGCAGCGCAGUGUUGUCAAUCGAUAAAUUAUCGAUUUU ..(((((...((((..(((.((..((.((((.((.(.(((((((............)))).....))).).)))))).))..)).)))..)))))))))(((((((...)))))))... ( -30.20) >DroSec_CAF1 1013 119 + 1 AGUGACACCCACACACGGCCGCACGGUACACACACACACACGCAAUCAAUACACACUGUGCACACUGUUUUUGGCGGCCCUGGCAGCGCAGUGUUGUCAAUCGAUAAGUUAUCGAUUUU .(((.....)))....(((((((((((...((((......................))))...))))).....))))))..((((((.....))))))((((((((...)))))))).. ( -30.65) >DroSim_CAF1 1210 119 + 1 AGUGACACCCACACACGGCCGCACGGUACACACACACACACGCAAUCAAUACACACUGUGCACACUGUUUUCGGCGGCCCUGGCAGCGCAGUGUUGUCAAUCGAUAAGUUAUCGAUUUU .(((.....)))....((((((.(((.(((....((((..................)))).....)))..)))))))))..((((((.....))))))((((((((...)))))))).. ( -31.57) >DroEre_CAF1 1091 117 + 1 AGUGACACUCACACACGGCCGCACGAAACGCACACA--CACACAAUCAAUACACACUGUGCACACUGUUUUCGGUGGCCCUGGCAACGCAGUGUUGCCAGUCGAUAAGUAAUCGAUUUU .(((.....)))....((((((.(((((.(((....--..((((............)))).....))))))))))))))((((((((.....))))))))(((((.....))))).... ( -37.60) >DroYak_CAF1 1272 117 + 1 AGUGACACUCACACACGGCCGCACGAAACGCACACA--CACACAAUCAAUACACACUGUGCACACUGUUUUCGGUGGCCCUGGCAACGCAGUGUUGCCAGCCGAUAAGUAAUCGAUUUU .(((.....)))....((((((.(((((.(((....--..((((............)))).....))))))))))))))((((((((.....)))))))).((((.....))))..... ( -36.10) >consensus AGUGACACCCACACACGGCCGCACGGUACGCACACACACACACAAUCAAUACACACUGUGCACACUGUUUUCGGCGGCCCUGGCAGCGCAGUGUUGUCAAUCGAUAAGUUAUCGAUUUU .(((.....)))....((((((.(((...(((......((((..............)))).....)))..))))))))).(((((((.....)))))))((((((.....))))))... (-29.14 = -27.82 + -1.32)

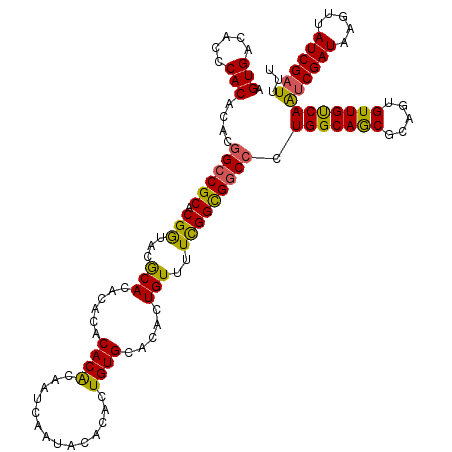
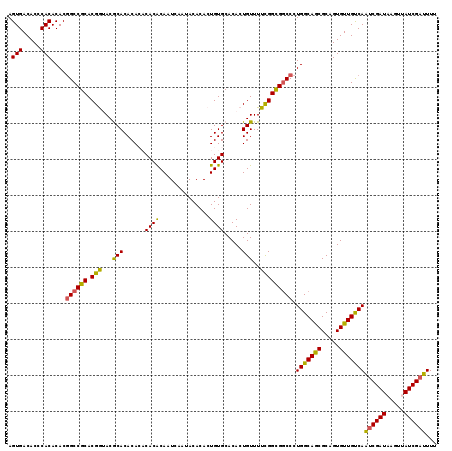
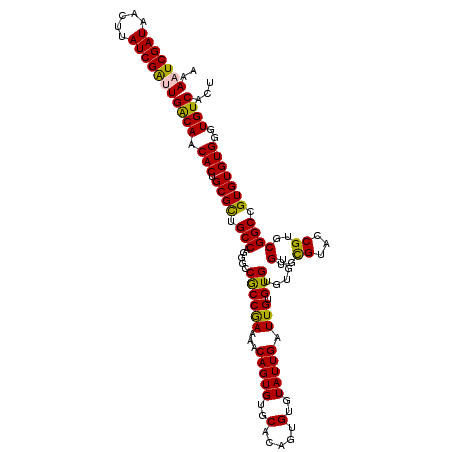
| Location | 14,961,096 – 14,961,215 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.09 |
| Mean single sequence MFE | -43.52 |
| Consensus MFE | -33.30 |
| Energy contribution | -32.34 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14961096 119 - 20766785 AAAAUCGAUAAUUUAUCGAUUGACAACACUGCGCUGCCAGGGACGCCAAAAACAGUGUGCACAGUGUGUAUUGAUUGUGUGUGUGUGUGCGUACCGUGCGGCGGUGUGUGGGUGUCACU ..((((((((...))))))))((((.(((.((((((((.((.((((((...(((...((((((((........))))))))))).)).)))).))....)))))))))))..))))... ( -45.10) >DroSec_CAF1 1013 119 - 1 AAAAUCGAUAACUUAUCGAUUGACAACACUGCGCUGCCAGGGCCGCCAAAAACAGUGUGCACAGUGUGUAUUGAUUGCGUGUGUGUGUGUGUACCGUGCGGCCGUGUGUGGGUGUCACU ..((((((((...))))))))((((...(..(((.((...((((((.....((.(((..(((((..((((.....))))..)...))))..))).)))))))))))))..).))))... ( -42.80) >DroSim_CAF1 1210 119 - 1 AAAAUCGAUAACUUAUCGAUUGACAACACUGCGCUGCCAGGGCCGCCGAAAACAGUGUGCACAGUGUGUAUUGAUUGCGUGUGUGUGUGUGUACCGUGCGGCCGUGUGUGGGUGUCACU ..((((((((...))))))))((((...(..(((.((...((((((((......(((..(((((..((((.....))))..)...))))..))))).)))))))))))..).))))... ( -43.00) >DroEre_CAF1 1091 117 - 1 AAAAUCGAUUACUUAUCGACUGGCAACACUGCGUUGCCAGGGCCACCGAAAACAGUGUGCACAGUGUGUAUUGAUUGUGUG--UGUGUGCGUUUCGUGCGGCCGUGUGUGAGUGUCACU ......(((.((((((...((((((((.....))))))))((((.((((((...(..((((((((........))))))))--..).....))))).).))))....)))))))))... ( -43.50) >DroYak_CAF1 1272 117 - 1 AAAAUCGAUUACUUAUCGGCUGGCAACACUGCGUUGCCAGGGCCACCGAAAACAGUGUGCACAGUGUGUAUUGAUUGUGUG--UGUGUGCGUUUCGUGCGGCCGUGUGUGAGUGUCACU ......(((.((((((...((((((((.....))))))))((((.((((((...(..((((((((........))))))))--..).....))))).).))))....)))))))))... ( -43.20) >consensus AAAAUCGAUAACUUAUCGAUUGACAACACUGCGCUGCCAGGGCCGCCGAAAACAGUGUGCACAGUGUGUAUUGAUUGUGUGUGUGUGUGCGUACCGUGCGGCCGUGUGUGGGUGUCACU ..(((((((.....)))))))((((.(((.((((.(((.....((((((...(((((..(.....)..))))).))).))).....(..((...))..)))).)))))))..))))... (-33.30 = -32.34 + -0.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:36 2006