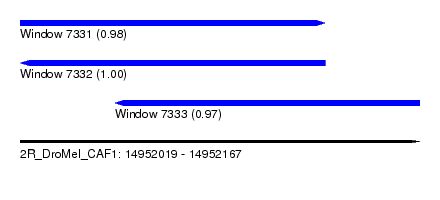
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,952,019 – 14,952,167 |
| Length | 148 |
| Max. P | 0.999937 |

| Location | 14,952,019 – 14,952,132 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 95.49 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -25.20 |
| Energy contribution | -26.04 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14952019 113 + 20766785 UGUGUCUGCCCGCUGUCUUUUAUCAGCCAGUAAUGGUUAUUUUUAAAAAUAUGAUUACUUAUUUCUCCGUAGUAGCCAGUGUGACCGAAUCCUUCUGCUGAUAAUAAGCCAGC ...........((((.(((((((((((......((((((((.....(((((........)))))......))))))))........((.....)).)))))))).))).)))) ( -24.00) >DroSec_CAF1 29348 113 + 1 UGUGCUUGCCCGCUGGCUUUUAUCAGCUAGUAAUGGGUAUUUUUAAAAAUAUGAUUACUUAUUUCUCCGUAGUAGCCAGUGUGACCGAAUCCUUCUGCUGAUAAUAAGCCACC ...((......))((((((((((((((.((....(((.((((......((((..(((((...........)))))...))))....))))))).)))))))))).)))))).. ( -25.80) >DroSim_CAF1 11509 113 + 1 UGUGCUUGCCCGCUGGCUUUUAUCAGCUAGUAAUGGUUAUUUUUAAAAAUAUGAUUACUUAUUUCUCCGUAGUAGCCAGUGUGACCGAAUCCUUCUGCUGAUAAUAAGCCAGC ...........((((((((((((((((......((((((((.....(((((........)))))......))))))))........((.....)).)))))))).)))))))) ( -31.50) >DroEre_CAF1 22618 113 + 1 UGUGCUUGCCCGCUGGCUUUUAUCAGCCAGUAAUGGUUAUUUUUAAAAAUAUGAUUACUUAUUUCUCCGUAUUAGCCAGUGUGACCGAAUCCUUAUGCUGAUAAUAGGCCAGC ...........((((((((((((((((.((...(((((((.......((((((..............)))))).......)))))))....))...)))))))).)))))))) ( -30.78) >DroYak_CAF1 11145 113 + 1 UAUGCUUGCCCGCUGGCUUUUAUCAGCCAGUAAUGGUUAUUUUUAAAAAUAUGAUUACUUAUUUCUCCGUAGUAGCCAGUGUGACCGAAUCCGUAUGCUGAUAAUAAGCCAAC ...((......))((((((((((((((......((((((((.....(((((........)))))......)))))))).......((....))...)))))))).)))))).. ( -26.90) >consensus UGUGCUUGCCCGCUGGCUUUUAUCAGCCAGUAAUGGUUAUUUUUAAAAAUAUGAUUACUUAUUUCUCCGUAGUAGCCAGUGUGACCGAAUCCUUCUGCUGAUAAUAAGCCAGC ...........((((((((((((((((......((((((((.....(((((........)))))......))))))))(..(......)..)....)))))))).)))))))) (-25.20 = -26.04 + 0.84)

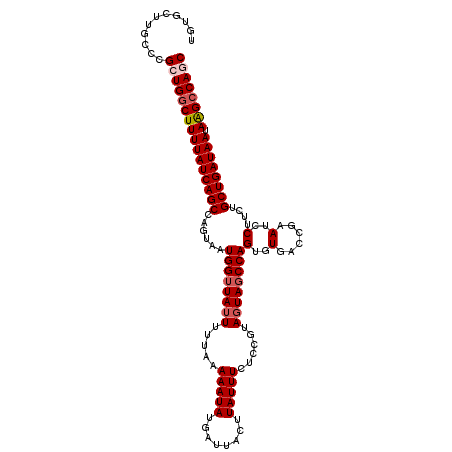
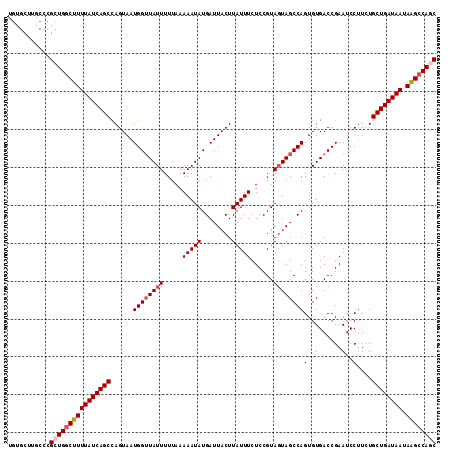
| Location | 14,952,019 – 14,952,132 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 95.49 |
| Mean single sequence MFE | -30.46 |
| Consensus MFE | -28.70 |
| Energy contribution | -29.38 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.68 |
| SVM RNA-class probability | 0.999937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14952019 113 - 20766785 GCUGGCUUAUUAUCAGCAGAAGGAUUCGGUCACACUGGCUACUACGGAGAAAUAAGUAAUCAUAUUUUUAAAAAUAACCAUUACUGGCUGAUAAAAGACAGCGGGCAGACACA ((((.(((.((((((((.......(((((((.....)))).(....).)))...((((((..(((((....)))))...)))))).))))))))))).))))........... ( -27.00) >DroSec_CAF1 29348 113 - 1 GGUGGCUUAUUAUCAGCAGAAGGAUUCGGUCACACUGGCUACUACGGAGAAAUAAGUAAUCAUAUUUUUAAAAAUACCCAUUACUAGCUGAUAAAAGCCAGCGGGCAAGCACA (.((((((.((((((((.......(((((((.....)))).(....).)))...((((((..(((((....)))))...)))))).)))))))))))))).)........... ( -29.80) >DroSim_CAF1 11509 113 - 1 GCUGGCUUAUUAUCAGCAGAAGGAUUCGGUCACACUGGCUACUACGGAGAAAUAAGUAAUCAUAUUUUUAAAAAUAACCAUUACUAGCUGAUAAAAGCCAGCGGGCAAGCACA ((((((((.((((((((.......(((((((.....)))).(....).)))...((((((..(((((....)))))...)))))).))))))))))))))))........... ( -34.30) >DroEre_CAF1 22618 113 - 1 GCUGGCCUAUUAUCAGCAUAAGGAUUCGGUCACACUGGCUAAUACGGAGAAAUAAGUAAUCAUAUUUUUAAAAAUAACCAUUACUGGCUGAUAAAAGCCAGCGGGCAAGCACA ((((((...((((((((..........((((.....))))..............((((((..(((((....)))))...)))))).))))))))..))))))........... ( -29.50) >DroYak_CAF1 11145 113 - 1 GUUGGCUUAUUAUCAGCAUACGGAUUCGGUCACACUGGCUACUACGGAGAAAUAAGUAAUCAUAUUUUUAAAAAUAACCAUUACUGGCUGAUAAAAGCCAGCGGGCAAGCAUA ((((((((.((((((((.......(((((((.....)))).(....).)))...((((((..(((((....)))))...)))))).))))))))))))))))........... ( -31.70) >consensus GCUGGCUUAUUAUCAGCAGAAGGAUUCGGUCACACUGGCUACUACGGAGAAAUAAGUAAUCAUAUUUUUAAAAAUAACCAUUACUGGCUGAUAAAAGCCAGCGGGCAAGCACA ((((((((.((((((((.......(((((((.....)))).(....).)))...((((((..(((((....)))))...)))))).))))))))))))))))........... (-28.70 = -29.38 + 0.68)

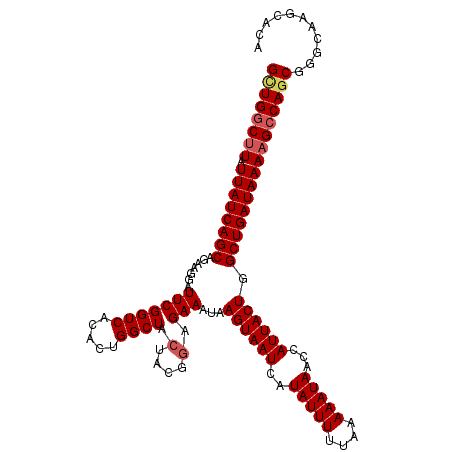
| Location | 14,952,054 – 14,952,167 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -28.76 |
| Consensus MFE | -24.06 |
| Energy contribution | -24.34 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14952054 113 - 20766785 CUCCAGUGAACCAGUUCCAAUUCGGGAACAGCCCUGCUGGCUUAUUAUCAGCAGAAGGAUUCGGUCACACUGGCUACUACGGAGAAAUAAGUAAUCAUAUUUUUAAAAAUAAC ..((((((.(((.(((((......)))))..(((((((((.......)))))))..))....)))..))))))...(....)(((((((.(....).)))))))......... ( -30.10) >DroSec_CAF1 29383 113 - 1 CUCCAGUGAACCAGUUCCAAUUCGGGAACAGCCCAGGUGGCUUAUUAUCAGCAGAAGGAUUCGGUCACACUGGCUACUACGGAGAAAUAAGUAAUCAUAUUUUUAAAAAUACC ((((.(((.....(((((......)))))((((.((((((((....(((........)))..)))))).))))))..)))))))............................. ( -28.40) >DroSim_CAF1 11544 113 - 1 CUCCAGUGAACCAGUUCCAAUUCGGGAACAGCCCAGCUGGCUUAUUAUCAGCAGAAGGAUUCGGUCACACUGGCUACUACGGAGAAAUAAGUAAUCAUAUUUUUAAAAAUAAC ((((((((..(((((.((((((((((.....))).(((((.......)))))....))))).))....))))).))))..))))............................. ( -27.40) >DroEre_CAF1 22653 113 - 1 AUCCAGUGAACCGGUUCCAAAUCGGGAACAGCCCAGCUGGCCUAUUAUCAGCAUAAGGAUUCGGUCACACUGGCUAAUACGGAGAAAUAAGUAAUCAUAUUUUUAAAAAUAAC ..((((((.((((..(((.....(((.....))).(((((.......)))))....)))..))))..))))))....(((..(....)..))).................... ( -31.30) >DroYak_CAF1 11180 113 - 1 CUCCAGCGAACCGGUUCCAAAUCGGGAACAGCCCAGUUGGCUUAUUAUCAGCAUACGGAUUCGGUCACACUGGCUACUACGGAGAAAUAAGUAAUCAUAUUUUUAAAAAUAAC ((((..((((((((((((......)))))((((.....)))).............))).))))(((.....)))......))))............................. ( -26.60) >consensus CUCCAGUGAACCAGUUCCAAUUCGGGAACAGCCCAGCUGGCUUAUUAUCAGCAGAAGGAUUCGGUCACACUGGCUACUACGGAGAAAUAAGUAAUCAUAUUUUUAAAAAUAAC ..((((((.(((...(((.....(((.....))).(((((.......)))))....)))...)))..)))))).........(((((((.(....).)))))))......... (-24.06 = -24.34 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:28 2006