| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,938,093 – 14,938,309 |
| Length | 216 |
| Max. P | 0.955108 |

| Location | 14,938,093 – 14,938,208 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.60 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -14.15 |
| Energy contribution | -13.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14938093 115 - 20766785 ACAGAUUAUUAUUCAGGGGUCAUAAAUGU-----CAUUUAAGAUCAAGAACAUAUUAUAUUUAAAUUUAUCCGGAAAUAUGAGCAAAUGUUUAAGGUGUAGCACCACAUUGACCCCCAUU ...............(((((((...((((-----.............((((((.((((((((............))))))))....))))))..((((...))))))))))))))).... ( -23.90) >DroSim_CAF1 24128 116 - 1 ACAGAUUAUUAUUCAGGGGUCAUGUAUGCA----CAUUUUAGAUAAAGAACAUAUGAUAUUUAAAUUAAUCCGAAAAUAUGAGCAAAUGUUUAAGGUGUAACACCACAUUGACCCCCAUU ...............((((((((((.((.(----(((((((((((..(..(((((.....................)))))..)...))))))))))))..))..))).))))))).... ( -24.90) >DroYak_CAF1 24304 116 - 1 UCAG-UUCUUAUUCAGGGGUUAUGUACAUAUAUGCUUUUUAGAUAAUACAAAAAUUAUAUUCCAAUUUUCUAUGAAAUUUCAGUA---GUUUUUGGGGUCACACCCUAUAGACCCCCCUU ....-..........((((((.((((......................((((((((((.....((((((....))))))...)))---)))))))(((.....))))))))))))).... ( -21.50) >consensus ACAGAUUAUUAUUCAGGGGUCAUGUAUGUA____CAUUUUAGAUAAAGAACAUAUUAUAUUUAAAUUUAUCCGGAAAUAUGAGCAAAUGUUUAAGGUGUAACACCACAUUGACCCCCAUU ...............((((((.....................................................((((((......))))))...(((......)))...)))))).... (-14.15 = -13.93 + -0.22)

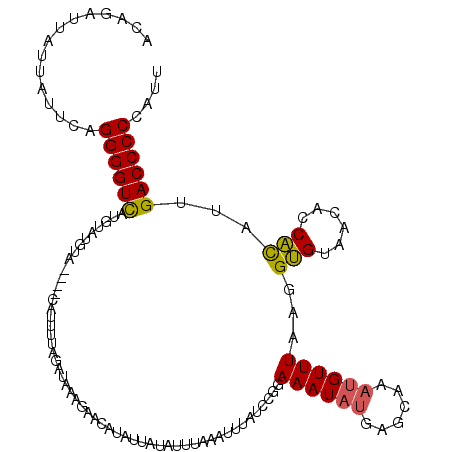

| Location | 14,938,208 – 14,938,309 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 92.41 |
| Mean single sequence MFE | -15.82 |
| Consensus MFE | -14.21 |
| Energy contribution | -13.99 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14938208 101 + 20766785 AGUUUCGCCACUGUCUAUGAAGCAUAACAACAAAAUAGACACACUACUCACUCACACACGCCGUGUUGCGUGAGCUUGACCAAACCCAAAAAACAAAAACA .((((.(.((.(((((((................))))))).....(((((.(((((.....))).)).)))))..)).).))))................ ( -17.29) >DroSim_CAF1 24244 94 + 1 AGUUCCGCCACUGUCUAUAAAGCAUAACAACAAAAUAGACACA-------CUCUCACACGCCGUGUUGCGUGAGCUUGACCAAACCCAAAAAACAAAAACA ((((((((...(((((((................)))))))..-------....(((.....)))..))).)))))......................... ( -14.39) >DroYak_CAF1 24420 101 + 1 AGUUCCGCCACUGUCUAUAAAGCAUAACAACAAAAUAGACACACUACACACUCACACACACCGUGUUGCGUGAGCUUGACCAAACCCAAAAAACAAAAACA ((((((((...(((((((................)))))))............((((.....)))).))).)))))......................... ( -15.79) >consensus AGUUCCGCCACUGUCUAUAAAGCAUAACAACAAAAUAGACACACUAC_CACUCACACACGCCGUGUUGCGUGAGCUUGACCAAACCCAAAAAACAAAAACA ((((((((...(((((((................))))))).............(((.....)))..))).)))))......................... (-14.21 = -13.99 + -0.22)

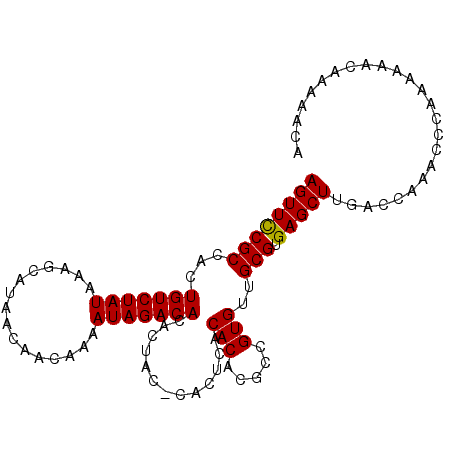
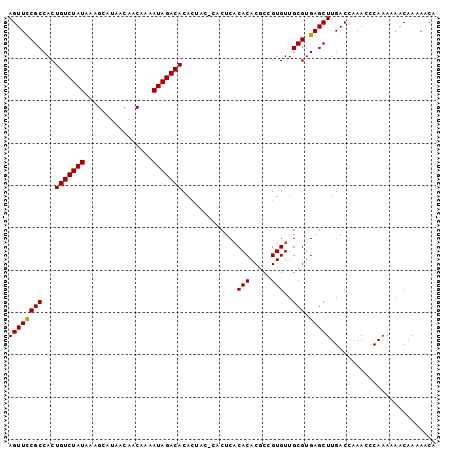
| Location | 14,938,208 – 14,938,309 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 92.41 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -26.79 |
| Energy contribution | -27.23 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14938208 101 - 20766785 UGUUUUUGUUUUUUGGGUUUGGUCAAGCUCACGCAACACGGCGUGUGUGAGUGAGUAGUGUGUCUAUUUUGUUGUUAUGCUUCAUAGACAGUGGCGAAACU ...............(((((.((((.((((((...((((.....))))..))))))....(((((((...((......))...))))))).)))).))))) ( -29.60) >DroSim_CAF1 24244 94 - 1 UGUUUUUGUUUUUUGGGUUUGGUCAAGCUCACGCAACACGGCGUGUGAGAG-------UGUGUCUAUUUUGUUGUUAUGCUUUAUAGACAGUGGCGGAACU ...............(((((.((((..(((((((........)))))))..-------..(((((((...((......))...))))))).)))).))))) ( -27.60) >DroYak_CAF1 24420 101 - 1 UGUUUUUGUUUUUUGGGUUUGGUCAAGCUCACGCAACACGGUGUGUGUGAGUGUGUAGUGUGUCUAUUUUGUUGUUAUGCUUUAUAGACAGUGGCGGAACU ...............(((((.((((.(((((((((........)))))))))........(((((((...((......))...))))))).)))).))))) ( -30.20) >consensus UGUUUUUGUUUUUUGGGUUUGGUCAAGCUCACGCAACACGGCGUGUGUGAGUG_GUAGUGUGUCUAUUUUGUUGUUAUGCUUUAUAGACAGUGGCGGAACU ...............(((((.((((.(((((((((........)))))))))........(((((((...((......))...))))))).)))).))))) (-26.79 = -27.23 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:21 2006