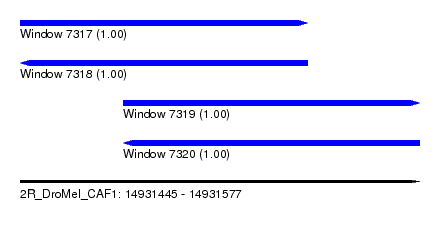
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,931,445 – 14,931,577 |
| Length | 132 |
| Max. P | 0.999933 |

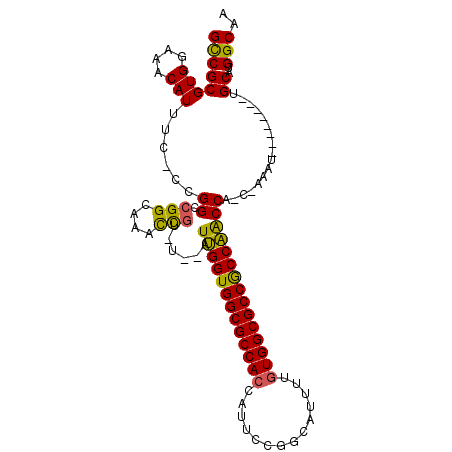
| Location | 14,931,445 – 14,931,540 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.32 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -33.32 |
| Energy contribution | -32.44 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.999114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
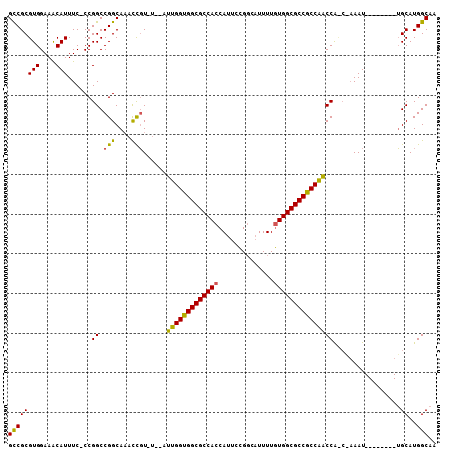
>2R_DroMel_CAF1 14931445 95 + 20766785 GCCGCGUGGAAACAUUUC-CCGGCCGGCAAAUCGU----AUUGGUGGCGCCACCAUUCCGGCAUUUUGUGGCGCCGCCAACCAUCAAAAU--------UGCAUGGCAA ((((((.((((....)))-)))..)))).......----.(((((((((((((..............)))))))))))))((((((....--------)).))))... ( -38.54) >DroYak_CAF1 17830 95 + 1 GCCGCGUGGAAGCAUUUC-CCGGCCGGCAAACCGUAUGUAUUGGUGGCGCCACCAUUCCGGCAUUUUGUGGCGCCGCCAACCA----AAU--------UGCAUGGCAA ((((((.((((....)))-)))..)))).....((((((((((((((((((((..............)))))))))))))...----...--------))))).)).. ( -37.94) >DroAna_CAF1 17627 105 + 1 GUCGCGUGGAAACAUUUCUCCGGCCGGCAUACUCUGU---UCGGUGGCGCCACCAUUCUGUCCUUUUAUGGCGCCACCGGCCGCCCAAAUAAUAGCCUGGCAUGGCAA ...(((((....)))......((((((((.....)))---).((((((((((................))))))))))))))))..........(((......))).. ( -39.99) >consensus GCCGCGUGGAAACAUUUC_CCGGCCGGCAAACCGU_U__AUUGGUGGCGCCACCAUUCCGGCAUUUUGUGGCGCCGCCAACCA_C_AAAU________UGCAUGGCAA ((((((((....)))......((.(((....)))......(((((((((((((..............))))))))))))))).................))..))).. (-33.32 = -32.44 + -0.88)

| Location | 14,931,445 – 14,931,540 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.32 |
| Mean single sequence MFE | -40.35 |
| Consensus MFE | -30.57 |
| Energy contribution | -30.47 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
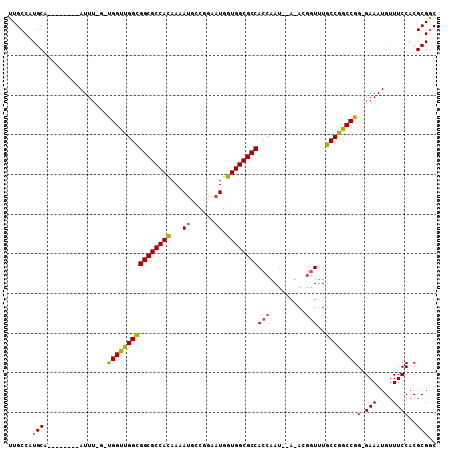
>2R_DroMel_CAF1 14931445 95 - 20766785 UUGCCAUGCA--------AUUUUGAUGGUUGGCGGCGCCACAAAAUGCCGGAAUGGUGGCGCCACCAAU----ACGAUUUGCCGGCCGG-GAAAUGUUUCCACGCGGC ..(((..(((--------(..(((...(((((.((((((((...((......)).)))))))).)))))----.))).))))..((..(-(((....))))..))))) ( -38.00) >DroYak_CAF1 17830 95 - 1 UUGCCAUGCA--------AUU----UGGUUGGCGGCGCCACAAAAUGCCGGAAUGGUGGCGCCACCAAUACAUACGGUUUGCCGGCCGG-GAAAUGCUUCCACGCGGC ..(((..((.--------...----((((((((((((((((...((......)).))))))))(((.........)))..))))))))(-(((....))))..))))) ( -39.80) >DroAna_CAF1 17627 105 - 1 UUGCCAUGCCAGGCUAUUAUUUGGGCGGCCGGUGGCGCCAUAAAAGGACAGAAUGGUGGCGCCACCGA---ACAGAGUAUGCCGGCCGGAGAAAUGUUUCCACGCGAC ((((...(((.(((...(((((....(..((((((((((((..............)))))))))))).---.).))))).)))))).(((((....)))))..)))). ( -43.24) >consensus UUGCCAUGCA________AUUU_G_UGGUUGGCGGCGCCACAAAAUGCCGGAAUGGUGGCGCCACCAAU__A_ACGGUUUGCCGGCCGG_GAAAUGUUUCCACGCGGC ......(((................((((((((((((((((...((......)).))))))))(((.........)))..))))))))(.(((....))))..))).. (-30.57 = -30.47 + -0.10)


| Location | 14,931,479 – 14,931,577 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.16 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -30.42 |
| Energy contribution | -29.87 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.64 |
| SVM RNA-class probability | 0.999933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14931479 98 + 20766785 -AUUGGUGGCGCCACCAUUCCGGCAUUUUGUGGCGCCGCCAACCAUCAAAAU--------UGCAUGGCAACACG---ACGAUAAUGAAAAUGCCGGCCACGAAAAGGAAA -.(((((((((((((..............)))))))))))))((.(((..((--------(((.((....)).)---.))))..)))......((....))....))... ( -33.34) >DroYak_CAF1 17867 95 + 1 UAUUGGUGGCGCCACCAUUCCGGCAUUUUGUGGCGCCGCCAACCA----AAU--------UGCAUGGCAACACG---AUGGUAAUGAAAAUGCCGGCCACAAAAAUGAAA ..(((((((((((((..............)))))))))))))((.----.((--------((..((....))))---))((((.(....))))))).............. ( -31.64) >DroAna_CAF1 17664 102 + 1 --UCGGUGGCGCCACCAUUCUGUCCUUUUAUGGCGCCACCGGCCGCCCAAAUAAUAGCCUGGCAUGGCAACACACGAAUGGUAAUGAAAAUGCCGGCAACGAAA------ --.(((((((((((................)))))))))))(((............(((......)))...........((((.(....)))))))).......------ ( -36.49) >consensus _AUUGGUGGCGCCACCAUUCCGGCAUUUUGUGGCGCCGCCAACCA_C_AAAU________UGCAUGGCAACACG___AUGGUAAUGAAAAUGCCGGCCACGAAAA_GAAA ..(((((((((((((..............)))))))))))))...................((.(((((.....................)))))))............. (-30.42 = -29.87 + -0.55)

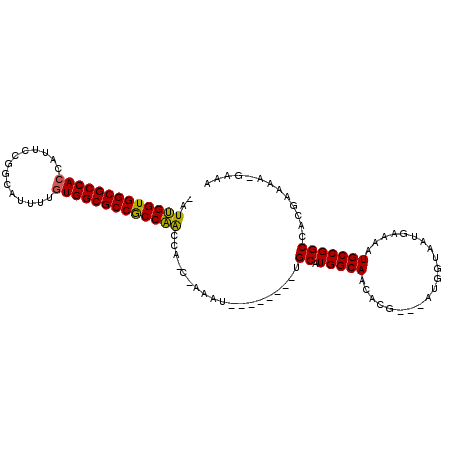
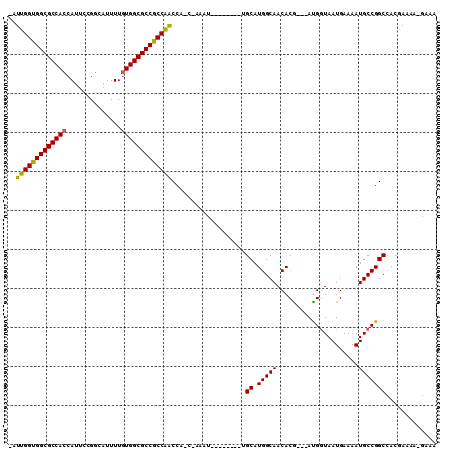
| Location | 14,931,479 – 14,931,577 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.16 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -28.44 |
| Energy contribution | -28.89 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.78 |
| SVM decision value | 4.03 |
| SVM RNA-class probability | 0.999766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14931479 98 - 20766785 UUUCCUUUUCGUGGCCGGCAUUUUCAUUAUCGU---CGUGUUGCCAUGCA--------AUUUUGAUGGUUGGCGGCGCCACAAAAUGCCGGAAUGGUGGCGCCACCAAU- ..............((((((((((......(((---(((...(((((.((--------....)))))))..))))))....))))))))))..(((((....)))))..- ( -35.70) >DroYak_CAF1 17867 95 - 1 UUUCAUUUUUGUGGCCGGCAUUUUCAUUACCAU---CGUGUUGCCAUGCA--------AUU----UGGUUGGCGGCGCCACAAAAUGCCGGAAUGGUGGCGCCACCAAUA ..((((....))))((((((((((.........---.(((((((((.((.--------...----..)))))))))))...))))))))))..(((((....)))))... ( -33.52) >DroAna_CAF1 17664 102 - 1 ------UUUCGUUGCCGGCAUUUUCAUUACCAUUCGUGUGUUGCCAUGCCAGGCUAUUAUUUGGGCGGCCGGUGGCGCCAUAAAAGGACAGAAUGGUGGCGCCACCGA-- ------....((((((((((....(((........)))...))))....((((......))))))))))((((((((((((..............)))))))))))).-- ( -40.04) >consensus UUUC_UUUUCGUGGCCGGCAUUUUCAUUACCAU___CGUGUUGCCAUGCA________AUUU_G_UGGUUGGCGGCGCCACAAAAUGCCGGAAUGGUGGCGCCACCAAU_ .........((((((.(((((................)))))))))))...................(((((.((((((((...((......)).)))))))).))))). (-28.44 = -28.89 + 0.45)

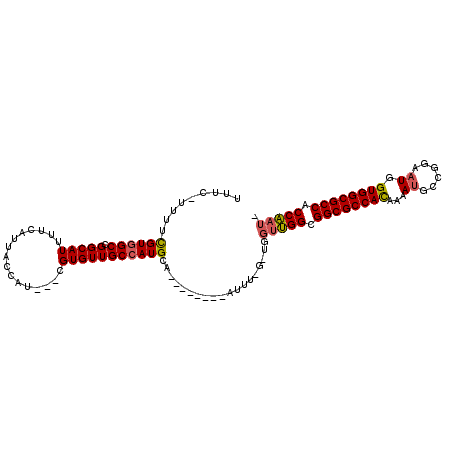
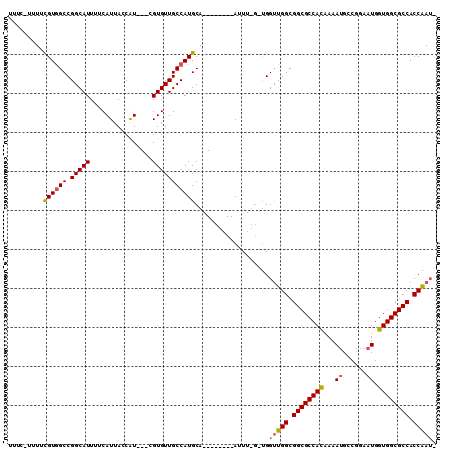
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:15 2006