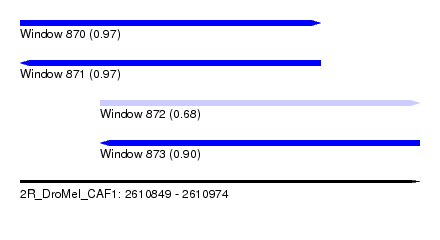
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,610,849 – 2,610,974 |
| Length | 125 |
| Max. P | 0.973212 |

| Location | 2,610,849 – 2,610,943 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -17.49 |
| Energy contribution | -18.08 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2610849 94 + 20766785 ACCGCGACAAUUGUAUGUGUCUCAACUGGCCAUCAAGUGGCCAAGUGGAGUUGGCCAAGAAAUGGCCAAAAGCCAAAGCUGAUGGGCGUGUAAU (((((((((........))))(((.((((((((...))))))...(((..(((((((.....)))))))...))).)).)))...))).))... ( -33.20) >DroSec_CAF1 45489 94 + 1 GCCGCGUCAAGUGUGUGUGUCUCAACUGGCCAUCGAGUGGCCAAGUGGAGUUGGCCAAGAAAUGGCCAAAAGCCAAAGCUGAUGGGCAUGUAAU (((.(((((.((...((.(((((.(((((((((...)))))).))).)))(((((((.....)))))))..))))..))))))))))....... ( -38.20) >DroSim_CAF1 42931 94 + 1 GCCGCGUCAAGUGUGUGUGUCUCAACUGGCCAUCGAGUGGCCAAGUGGAGUUGGCCAAGAAAUGGCCAAAAGCCAAAGCUGAUGGGCGUGUAAU (((.(((((.((...((.(((((.(((((((((...)))))).))).)))(((((((.....)))))))..))))..))))))))))....... ( -38.20) >DroEre_CAF1 44763 85 + 1 GCGGCGACAAGUG----UCUCGCUACUGGCCCUCAAGUGGCCGA-----CUUGGCCAAGAAAUGGCCAGCAGCCAAAGCUGAUGGGCGUGUAAU ((((.(((....)----))))))(((..(((((((..((((.(.-----((.(((((.....)))))))).))))....))).))))..))).. ( -35.00) >DroWil_CAF1 60461 71 + 1 ------CCAAGUG-----GUUUCAACUGACAAUCAAGUGGCU---UGUAGUUAG-------AGGGCCA--AGGCAAGGUUGAUGGGCGUCUAAU ------(((..((-----(((((....)).)))))..)))..---....(((((-------(.(.(((--.(((...)))..))).).)))))) ( -18.30) >DroYak_CAF1 43177 85 + 1 GCCACGACAAGUG----CGUCUCUACUGGCUCUCAAGUGGCCAA-----CUUGGUUAGGAAAUGGCCAAAAGCCAAAGCUGAUGGGCGUGUAAU ......(((.((.----((((.....(((((......((((((.-----((.....))....))))))..))))).....)))).)).)))... ( -24.90) >consensus GCCGCGACAAGUG____UGUCUCAACUGGCCAUCAAGUGGCCAA_UGGAGUUGGCCAAGAAAUGGCCAAAAGCCAAAGCUGAUGGGCGUGUAAU ..........................((.((((((..(((((((......))))))).....((((.....))))....)))))).))...... (-17.49 = -18.08 + 0.59)

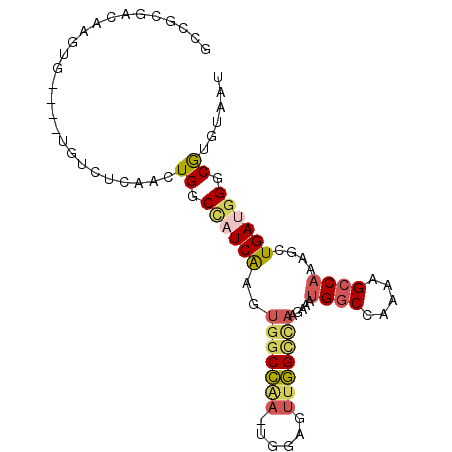
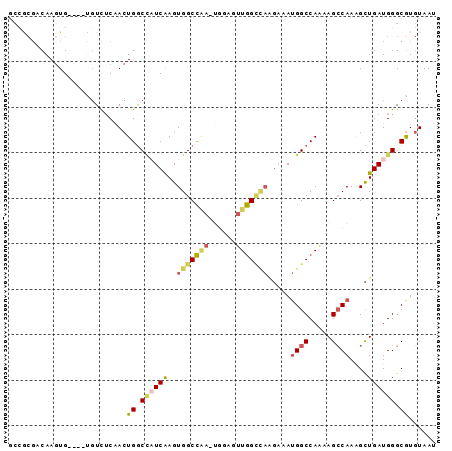
| Location | 2,610,849 – 2,610,943 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -13.93 |
| Energy contribution | -15.85 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2610849 94 - 20766785 AUUACACGCCCAUCAGCUUUGGCUUUUGGCCAUUUCUUGGCCAACUCCACUUGGCCACUUGAUGGCCAGUUGAGACACAUACAAUUGUCGCGGU ......(((...(((((..(((...(((((((.....)))))))..)))..((((((.....)))))))))))((((........))))))).. ( -34.70) >DroSec_CAF1 45489 94 - 1 AUUACAUGCCCAUCAGCUUUGGCUUUUGGCCAUUUCUUGGCCAACUCCACUUGGCCACUCGAUGGCCAGUUGAGACACACACACUUGACGCGGC .......((((...(((....)))..((((((((...(((((((......)))))))...))))))))((..((.........))..))).))) ( -32.20) >DroSim_CAF1 42931 94 - 1 AUUACACGCCCAUCAGCUUUGGCUUUUGGCCAUUUCUUGGCCAACUCCACUUGGCCACUCGAUGGCCAGUUGAGACACACACACUUGACGCGGC ......(((.....(((....)))..((((((((...(((((((......)))))))...))))))))((..((.........))..))))).. ( -32.70) >DroEre_CAF1 44763 85 - 1 AUUACACGCCCAUCAGCUUUGGCUGCUGGCCAUUUCUUGGCCAAG-----UCGGCCACUUGAGGGCCAGUAGCGAGA----CACUUGUCGCCGC ..(((..((((.((((...(((((((((((((.....))))).))-----.)))))).))))))))..)))(((.((----(....)))..))) ( -37.30) >DroWil_CAF1 60461 71 - 1 AUUAGACGCCCAUCAACCUUGCCU--UGGCCCU-------CUAACUACA---AGCCACUUGAUUGUCAGUUGAAAC-----CACUUGG------ ....((((...(((((...((.((--((.....-------.......))---)).)).))))))))).........-----.......------ ( -8.60) >DroYak_CAF1 43177 85 - 1 AUUACACGCCCAUCAGCUUUGGCUUUUGGCCAUUUCCUAACCAAG-----UUGGCCACUUGAGAGCCAGUAGAGACG----CACUUGUCGUGGC .......((((.....((((((((((((((((.............-----.))))))....)))))))).)).((((----....))))).))) ( -24.54) >consensus AUUACACGCCCAUCAGCUUUGGCUUUUGGCCAUUUCUUGGCCAACUCCA_UUGGCCACUUGAUGGCCAGUUGAGACA____CACUUGUCGCGGC .......(.(((((((...(((((.(((((((.....)))))))........))))).))))))).)........................... (-13.93 = -15.85 + 1.92)

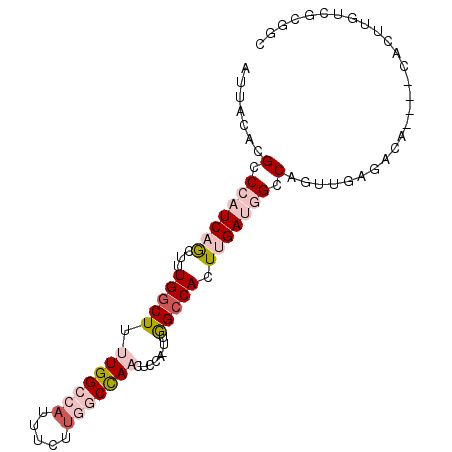
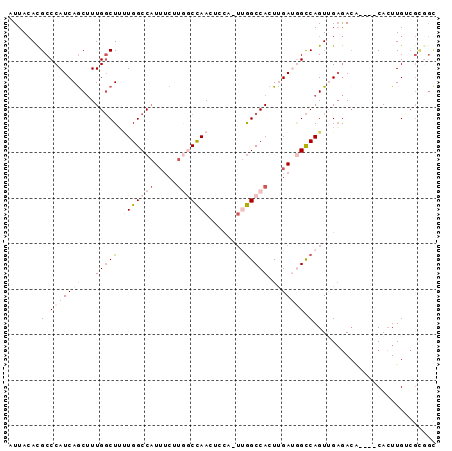
| Location | 2,610,874 – 2,610,974 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -21.71 |
| Energy contribution | -22.43 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2610874 100 + 20766785 CUGGCCAUCAAGUGGCCAAGUGGAGUUGGCCAAGAAAUGGCCAAAAGCCAAAGCUGAUGGGCGUGUAAUUGAUUCGGAGGCACAUGAUGGUGUGGGUGGU- ...(((((((.(((.((...(((..(((((((.....)))))))...)))...((((....((......))..)))).))))).))))))).........- ( -34.80) >DroSec_CAF1 45514 100 + 1 CUGGCCAUCGAGUGGCCAAGUGGAGUUGGCCAAGAAAUGGCCAAAAGCCAAAGCUGAUGGGCAUGUAAUUGAUUCGG-GGCACAUGAUGGUGUGGGUGGUG ...(((((((.(((.((...(((..(((((((.....)))))))...)))..(((....)))..............)-).))).))))))).......... ( -36.10) >DroSim_CAF1 42956 100 + 1 CUGGCCAUCGAGUGGCCAAGUGGAGUUGGCCAAGAAAUGGCCAAAAGCCAAAGCUGAUGGGCGUGUAAUUGAUUCGGGGGCACAUGAUGGUGUGGGUGGU- ...(((((((.(((.((...(((..(((((((.....)))))))...)))...((((....((......))..)))))).))).))))))).........- ( -36.60) >DroEre_CAF1 44784 93 + 1 CUGGCCCUCAAGUGGCCGA-----CUUGGCCAAGAAAUGGCCAGCAGCCAAAGCUGAUGGGCGUGUAAUUGAUUCGG--GGAUGUCAUGGUGUGGGUGGU- ...((((.((.(((((...-----(((((.(((...(((.(((.((((....)))).))).)))....)))..))))--)...)))))..)).))))...- ( -32.60) >DroYak_CAF1 43198 94 + 1 CUGGCUCUCAAGUGGCCAA-----CUUGGUUAGGAAAUGGCCAAAAGCCAAAGCUGAUGGGCGUGUAAUUGAUUCGG-GGGACAUAAUGAUGUAGGUGGU- .(((((.......))))).-----(((((((((...(((.(((..(((....)))..))).)))....)))).))))-)..((((....)))).......- ( -21.80) >DroAna_CAF1 44454 93 + 1 UUGGCCGUCAAGUGGCCAAGU----UAGACCAACAAUAGCCCAAAAGCCAAAGCUGAUGGGCGUCUAAUUGUUACAGAG---UUUGAUGGGCGGGGUG-UG (((.((((((((((((..(((----(((((........(((((..(((....)))..))))))))))))))))))....---.))))))).)))....-.. ( -31.70) >consensus CUGGCCAUCAAGUGGCCAAGU___GUUGGCCAAGAAAUGGCCAAAAGCCAAAGCUGAUGGGCGUGUAAUUGAUUCGG_GGCACAUGAUGGUGUGGGUGGU_ ...(((((((..(((((((......)))))))....(((.(((..(((....)))..))).)))....................))))))).......... (-21.71 = -22.43 + 0.73)

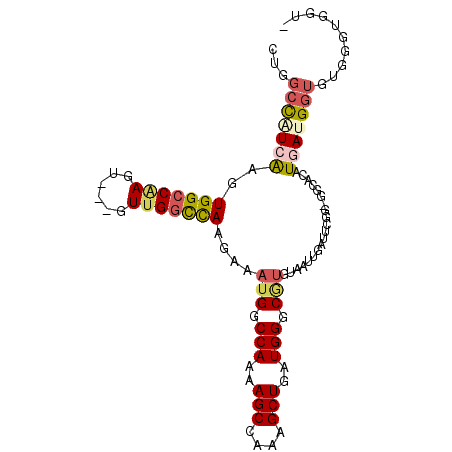
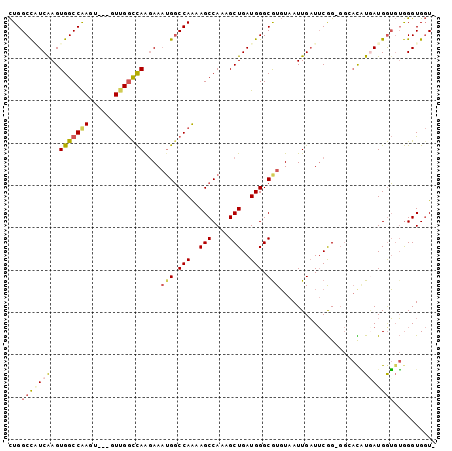
| Location | 2,610,874 – 2,610,974 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -26.71 |
| Consensus MFE | -16.95 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2610874 100 - 20766785 -ACCACCCACACCAUCAUGUGCCUCCGAAUCAAUUACACGCCCAUCAGCUUUGGCUUUUGGCCAUUUCUUGGCCAACUCCACUUGGCCACUUGAUGGCCAG -......((((......))))..................(((.(......).)))...((((((((...(((((((......)))))))...)))))))). ( -28.90) >DroSec_CAF1 45514 100 - 1 CACCACCCACACCAUCAUGUGCC-CCGAAUCAAUUACAUGCCCAUCAGCUUUGGCUUUUGGCCAUUUCUUGGCCAACUCCACUUGGCCACUCGAUGGCCAG .......((((......))))..-...............(((.(......).)))...((((((((...(((((((......)))))))...)))))))). ( -27.90) >DroSim_CAF1 42956 100 - 1 -ACCACCCACACCAUCAUGUGCCCCCGAAUCAAUUACACGCCCAUCAGCUUUGGCUUUUGGCCAUUUCUUGGCCAACUCCACUUGGCCACUCGAUGGCCAG -......((((......))))..................(((.(......).)))...((((((((...(((((((......)))))))...)))))))). ( -28.20) >DroEre_CAF1 44784 93 - 1 -ACCACCCACACCAUGACAUCC--CCGAAUCAAUUACACGCCCAUCAGCUUUGGCUGCUGGCCAUUUCUUGGCCAAG-----UCGGCCACUUGAGGGCCAG -.....................--...............((((.((((...(((((((((((((.....))))).))-----.)))))).))))))))... ( -30.30) >DroYak_CAF1 43198 94 - 1 -ACCACCUACAUCAUUAUGUCCC-CCGAAUCAAUUACACGCCCAUCAGCUUUGGCUUUUGGCCAUUUCCUAACCAAG-----UUGGCCACUUGAGAGCCAG -......................-...............((......))..(((((((((((((.............-----.))))))....))))))). ( -17.64) >DroAna_CAF1 44454 93 - 1 CA-CACCCCGCCCAUCAAA---CUCUGUAACAAUUAGACGCCCAUCAGCUUUGGCUUUUGGGCUAUUGUUGGUCUA----ACUUGGCCACUUGACGGCCAA ..-......(((..((((.---......((((((.....(((((..(((....)))..))))).))))))((((..----....))))..)))).)))... ( -27.30) >consensus _ACCACCCACACCAUCAUGUGCC_CCGAAUCAAUUACACGCCCAUCAGCUUUGGCUUUUGGCCAUUUCUUGGCCAAC___ACUUGGCCACUUGAUGGCCAG .......................................(((.(......).)))..((((((.((...(((((((......)))))))...)).)))))) (-16.95 = -17.70 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:03 2006