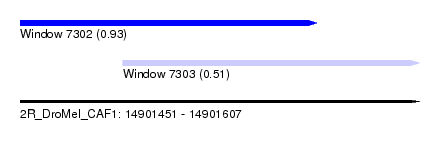
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,901,451 – 14,901,607 |
| Length | 156 |
| Max. P | 0.930287 |

| Location | 14,901,451 – 14,901,567 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.40 |
| Mean single sequence MFE | -38.03 |
| Consensus MFE | -26.42 |
| Energy contribution | -25.98 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
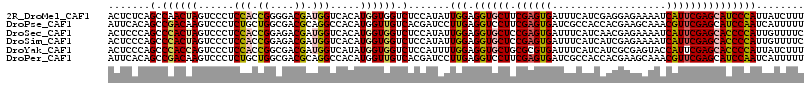
>2R_DroMel_CAF1 14901451 116 + 20766785 ACUCUCAGCCAACUAGUCCCUCCACCGGGGACGAUGGUCACAUGGUGGUCUCCAUAUUGGAGGUGCUUCGAGUGAUUUCAUCGAGGAGAAAAUCAUUCGAGCAUCCCAUUAUCUUU .......((((....((((((.....))))))..))))...(((((((.((((.....))))(((((.((((((((((..((...))..))))))))))))))).))))))).... ( -41.40) >DroPse_CAF1 83966 116 + 1 AUUCACAGCCGACAAGUCCCUCUGCUGGCGACGCAGGCCACAUGGUUGUCACGAUCCUUGAGGUCCUUCGAGUGAUCGCCACCACGAAGCAAACGUUCGAGCAUCCAAUCAUUUUU .......(((((...((.(.((((.((((((((...(((....)))))))..((((((((((....)))))).)))))))).)).)).)...))..))).)).............. ( -29.80) >DroSec_CAF1 73502 116 + 1 ACUCCCAGCCCACUAGUCCCUCCACCGGAGACGAUGGUCACAUGGUGGUCUCCAUAUUGGAGGUGCUCCGAGUGAUUUCAUCAACGAGAAAAUCAUUCGAGCACCCCAUUGUUUUC ......((.((((((......(((.((....)).))).....)))))).))..(((.(((.(((((((.(((((((((..((...))..))))))))))))))))))).))).... ( -44.30) >DroSim_CAF1 75634 116 + 1 ACUCCCAGCCCACUAGUCCCUCCACCGGAGACGAUGGUCACAUGGUGGUCUCCAUAUUGGAGGUGCUCCGAGUGAUUUCAUCAUCGAGAAAAUCAUUCGAGCACCCCAUUGUUUUC ......((.((((((......(((.((....)).))).....)))))).))..(((.(((.(((((((.(((((((((..((...))..))))))))))))))))))).))).... ( -44.30) >DroYak_CAF1 75415 116 + 1 ACUCCCAGCCCACCAGUCCCUCCACCGGCGACGAUGGUCAUAUGGUGGUCUCCAUUUUGGAGGUGCUGCGCGUGAUUUCAUCAUCGCGAGUACCAUUCGAGCACCCCAUUAUCUUU .......((((((((......(((.((....)).))).....))))))........(((((((((((.((((((((...)))).)))))))))).))))))).............. ( -38.60) >DroPer_CAF1 80964 116 + 1 AUUCACAGCCGACAAGUCCCUCUGCUGGCGACGCAGGCCACAUGGUUGUCACGAUCCUUGAGGUCCUUCGAGUGAUCGCCACCACGAAGCAAACGUUCGAGCAUCCAAUCAUUUUU .......(((((...((.(.((((.((((((((...(((....)))))))..((((((((((....)))))).)))))))).)).)).)...))..))).)).............. ( -29.80) >consensus ACUCCCAGCCCACUAGUCCCUCCACCGGCGACGAUGGUCACAUGGUGGUCUCCAUAUUGGAGGUGCUUCGAGUGAUUUCAUCAACGAGAAAAUCAUUCGAGCACCCCAUUAUUUUU .......(.((((((......(((.((....)).))).....)))))).).......(((.((((((.((((((...................)))))))))))))))........ (-26.42 = -25.98 + -0.44)
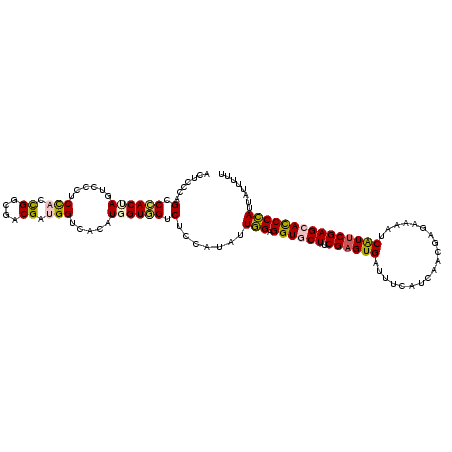
| Location | 14,901,491 – 14,901,607 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.99 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -22.99 |
| Energy contribution | -23.35 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14901491 116 + 20766785 CAUGGUGGUCUCCAUAUUGGAGGUGCUUCGAGUGAUUUCAUCGAGGAGAAAAUCAUUCGAGCAUCCCAUUAUCUUUUUGAUCAACGGAUCCGAGGAAAAUUCGCUGCAGGCAUCUC .((((......))))....(((((((((((((((((((..((...))..)))))))))))(((.(..(((.(((((..((((....)))).))))).)))..).))).)))))))) ( -35.60) >DroSec_CAF1 73542 115 + 1 CAUGGUGGUCUCCAUAUUGGAGGUGCUCCGAGUGAUUUCAUCAACGAGAAAAUCAUUCGAGCACCCCAUUGUUUUCUUAAUCAAUGGAUCCGAG-AAAAUUCGCUGCAGGCAUCUC .((((......)))).((((((((((((.(((((((((..((...))..))))))))))))))))((((((.((....)).)))))).))))).-.......((.....))..... ( -38.20) >DroSim_CAF1 75674 116 + 1 CAUGGUGGUCUCCAUAUUGGAGGUGCUCCGAGUGAUUUCAUCAUCGAGAAAAUCAUUCGAGCACCCCAUUGUUUUCUUAAUCAAUGGAUCCGAGGAAAAUUCGCUGCAGGCAUCUC ...(((((..(((...((((((((((((.(((((((((..((...))..))))))))))))))))((((((.((....)).)))))).))))))))....)))))........... ( -39.10) >DroEre_CAF1 74298 116 + 1 UAUGGUGGUCUCCAUAUUGGAGGUGCUGCGAGUGAUUUCGUCGUCGAGAAUUCCAUUCGAGCACCCCAUUAUCUUUCUGAUCAACGGAUCCGAGGAAAAUUCACUGCAGGCAUCGC ...(((((..((((((.(((.((((((.((((((.(((((....)))))....))))))))))))))).)))...((.((((....)))).)))))....)))))........... ( -37.00) >DroYak_CAF1 75455 116 + 1 UAUGGUGGUCUCCAUUUUGGAGGUGCUGCGCGUGAUUUCAUCAUCGCGAGUACCAUUCGAGCACCCCAUUAUCUUUUUGAUCAACGGAUCCGAGGAGAACUCACUGCAGGCAUCUC ...((((.(((((.....)))((((((.((((((((...)))).))))))))))....)).))))......(((((((((((....))).)))))))).................. ( -36.70) >DroPer_CAF1 81004 116 + 1 CAUGGUUGUCACGAUCCUUGAGGUCCUUCGAGUGAUCGCCACCACGAAGCAAACGUUCGAGCAUCCAAUCAUUUUUCUUAUUAACGGCUCCGAGGAGAACUCUUUGCAGGCCUCGC .(((((((...(((((((((((....)))))).)))))......((((.......))))......))))))).............((((.((((((....))))))..)))).... ( -29.60) >consensus CAUGGUGGUCUCCAUAUUGGAGGUGCUCCGAGUGAUUUCAUCAACGAGAAAAUCAUUCGAGCACCCCAUUAUCUUUUUGAUCAACGGAUCCGAGGAAAAUUCGCUGCAGGCAUCUC ...(((.((((.((...(((.((((((.((((((..(((........)))...))))))))))))))).......................(((.....)))..)).))))))).. (-22.99 = -23.35 + 0.36)

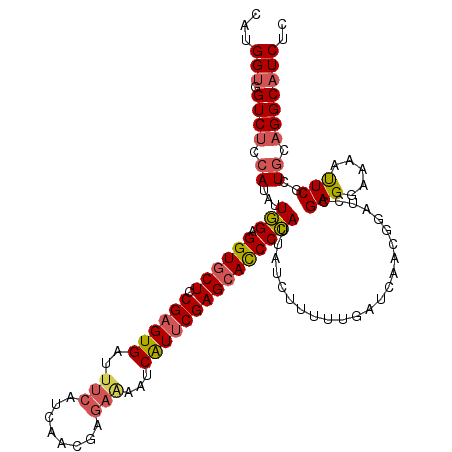
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:00 2006