
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,875,593 – 14,875,742 |
| Length | 149 |
| Max. P | 0.716251 |

| Location | 14,875,593 – 14,875,702 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 75.60 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -16.27 |
| Energy contribution | -16.38 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
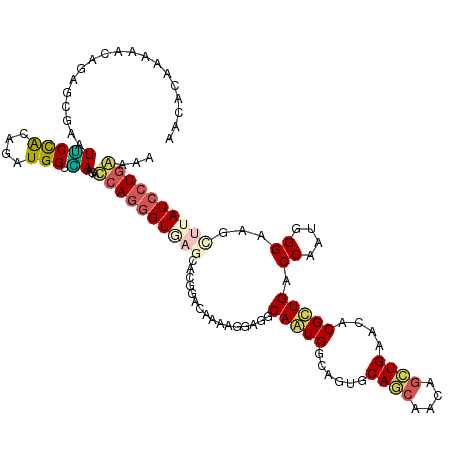
>2R_DroMel_CAF1 14875593 109 - 20766785 GGGUGAGAACAGAUAAAAGUAGGCAGUGGCAGUGCAGCAAAAGCUGAACCCGCUGACCAAUGGGAAGCUUAUCCUGAAAAAAGAUUUAGCUUUAGUCAACCAAUAAAUC ((((....)).(((.(((((.((((((((..((.((((....)))).)))))))).))...((((......)))).............))))).)))..))........ ( -29.30) >DroPse_CAF1 54166 103 - 1 UGGUGAAGAUGGCCAGGAAGAGGCAGUGACAGUGCAGCAGCAUCUGGACCCGCUGUCCAAUGGGCAGUUUGUCCUGGAAAAG-AAAUAAA----AUCAA-AUAUGAAUA ((((........((((((...((..((..((((((....))).))).))))(((((((...)))))))...)))))).....-.......----)))).-......... ( -25.85) >DroSim_CAF1 49655 108 - 1 GGGUAAGAACGGAUAAGAGGAUGCAGUGGCAGUGCAGCAAAAGCUGAACCCGCUGACCAAUGGGAAGCUUAUCCUGAAAAA-AAUGUAGCGUUAGUCAUACAAUAAAUC ..........(((((((....(.((.((((((((((((....))))....))))).))).)).)...))))))).......-..((((((....))..))))....... ( -26.20) >DroEre_CAF1 48169 108 - 1 GGGCAAGAACGGAUAAAAGCACGCAGUGGCAGUGCAGCAAAAGCUGAAUCCGCUGGCCAAUGGGAAGCUUAUCCUGGAAUAA-AAAUAAUGUUAGUUAAACAGUGAAAC ..((......(((((..(((.(.((.((((((((((((....))))....)))).)))).)).)..))))))))........-......((((.....))))))..... ( -25.20) >DroYak_CAF1 49488 109 - 1 GGGUGAGGACGGACAGAAGCACGCAGUGGCAGUGCAGCAAAAGCUGAAUCCGCUGGCCAAUGGGAAGCUUAUCCUGAAAAGGGAUUUAAAUUUAGUCAAAAGAUUAAAC .(((.((..((((.....((((((....)).))))(((....)))...)))))).))).............((((....)))).......(((((((....))))))). ( -28.80) >DroPer_CAF1 51012 103 - 1 UGGUGAAGAUGGCCAGGAAGAGGCAGUGACAGUGCAGCAGCAUCUGGACCCGCUGUCCAAUGGGCAGUUUGUCCUGGAAAAG-AAAUAAA----AUCAA-AGAUGAAUA ((((........((((((...((..((..((((((....))).))).))))(((((((...)))))))...)))))).....-.......----)))).-......... ( -25.85) >consensus GGGUGAGAACGGACAAAAGGAGGCAGUGGCAGUGCAGCAAAAGCUGAACCCGCUGACCAAUGGGAAGCUUAUCCUGAAAAAG_AAAUAAA_UUAGUCAAAAAAUGAAUC (((((((................((((((..((.((((....)))).)))))))).((....))...)))))))................................... (-16.27 = -16.38 + 0.11)



| Location | 14,875,622 – 14,875,742 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.83 |
| Mean single sequence MFE | -35.18 |
| Consensus MFE | -16.71 |
| Energy contribution | -16.77 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14875622 120 - 20766785 AAAACAAAAACAGAGCGAAUUCCACAGAUGGUGAAGAUCAGGGUGAGAACAGAUAAAAGUAGGCAGUGGCAGUGCAGCAAAAGCUGAACCCGCUGACCAAUGGGAAGCUUAUCCUGAAAA ......................(((.....)))....((((((((((.((........)).((((((((..((.((((....)))).)))))))).)).........))))))))))... ( -32.60) >DroGri_CAF1 51911 120 - 1 AGCACAAAUGCAGAGCGAAUACCACAGGUGGUUAGGACCAGAGUGAGCAGUGCCAGAAGCAGGCAAUGGCAAUGCAGCAGCAGCUGUACACGCUGCCCAAUGGGGAAGCGAUCCUGUGGA .(((....)))..........(((((((((((....))))..(((.((..((((.......))))...))..((((((....)))))))))(((.(((....))).)))...))))))). ( -45.50) >DroSec_CAF1 47451 118 - 1 AGGACAAAAACAGAGCGAAUUCCGCAGAUGGUAAAGAUCAGGGUAAGAACGGAUAAGAGGAUGCAGUGGCAGUGCAGCAA-AGCUGAACCCGCUGACCAAUGGGAAGCUUAUC-UGAAAA ..........((((.....((((.((..((((....(((.(........).))).........((((((..((.((((..-.)))).)))))))))))).)))))).....))-)).... ( -29.20) >DroWil_CAF1 81512 120 - 1 AACACGAAAGCCGAACGAAUGCCACAGAUGGUCAAGACCAGGGUAAGCAAGGCCAAGAAGAGGCAAUGCGAAUGCAACAACAAUUGCACACGUUGGCCAAGUGGAAGUUUAUCCUAGAAC ..(((....(((.((((..((((.....((((....)))).)))).(((..(((.......)))..)))...(((((......)))))..)))))))...)))................. ( -31.90) >DroYak_CAF1 49517 120 - 1 AGGACAAAAACAGAUCGAAUUCCACAGAUGGUGAAGAUCAGGGUGAGGACGGACAGAAGCACGCAGUGGCAGUGCAGCAAAAGCUGAAUCCGCUGGCCAAUGGGAAGCUUAUCCUGAAAA .(..(..(..(.((((.....(((....)))....)))).)..)..)..)(((...((((.(.((.((((((((((((....))))....)))).)))).)).)..)))).)))...... ( -31.60) >DroMoj_CAF1 73795 120 - 1 AACACAAAUGCCGAGCGGAUGCUGCAGAUGGUCAGCACCAGGGUGAGCAGCGCCAGGAAGAGGCAAUGGCAGUGCAACAACAGCUGCACACGUUGUCCAAUGGGCAGGCGAUCCUGGAGA ..........((....)).((((((.....).)))))(((((((..(((((((((...........)))).((......)).)))))...((((((((...))))).))))))))))... ( -40.30) >consensus AACACAAAAACAGAGCGAAUUCCACAGAUGGUCAAGACCAGGGUGAGCACGGACAAAAGGAGGCAAUGGCAGUGCAGCAACAGCUGAACACGCUGACCAAUGGGAAGCUUAUCCUGAAAA ...................(((((....))).))...((((((((((................(((((......((((....))))....))))).((....))...))))))))))... (-16.71 = -16.77 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:48 2006