| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,864,333 – 14,864,438 |
| Length | 105 |
| Max. P | 0.692245 |

| Location | 14,864,333 – 14,864,438 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.24 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -23.69 |
| Energy contribution | -23.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14864333 105 - 20766785 AAGACAAUACGGAGGGUCUACGGAGUGCUUUGGCCCGUUUACGCAACAGAACGGGCCCCUCUCUUCCA------CAAC---AACC------CACCCCGCCGGCCUCUCCCCUGCCCCUUG ..........((((((((..(((.(((....(((((((((........)))))))))...........------....---....------))).)))..)))).))))........... ( -32.80) >DroVir_CAF1 42517 105 - 1 AAGACAAUACGGAGGGUUUACGGAGUGCUUUGGCCCGUUUACGCAACAGAACGGGCCCCUCAAUUCCA------CAACCGCAACC------CACUCCGCCAGCCUCACCAUUGCCCU--- ....((((...((((.(...(((((((....(((((((((........)))))))))...........------(....).....------)))))))..).))))...))))....--- ( -32.40) >DroGri_CAF1 39390 105 - 1 AAGACAAUACGGAGGGUUUACGGAGUGCAUUGGCCCGUUUACGCAACAGAACGGGCCCCUCAAUUCCA------CAGCCGCAACC------CACUCCGCCAGCCUCACCAUUGCCCA--- ....((((...((((.(...(((((((....(((((((((........)))))))))...........------(....).....------)))))))..).))))...))))....--- ( -32.40) >DroWil_CAF1 62337 114 - 1 AAGACAAUACGGAGGGCUUACGAAGUGCUUUGGCCCGUUUACGCAACAGAACGGGCCCCUCAAUUCCAACAACCCAAC---AACCUACCACCACACCGCCAGCCUCCCCC-U--CCCUAA ..........((((((.....(((.((....(((((((((........)))))))))...)).)))............---................(....)....)))-)--)).... ( -26.60) >DroMoj_CAF1 59557 104 - 1 AAGACAAUACGGAGGGUUUACGGAGUGCUUUGGCCCGUUUACGCAACAGAACGGGCCCCUCAAUUCCA------CAGCCGCA-CC------CACUCCGCCCGCCUCGCCAUUGCCCA--- ..........((.((((...(((((((....(((((((((........)))))))))...........------(....)..-..------)))))))...))))..))........--- ( -31.30) >DroPer_CAF1 37603 105 - 1 AAGACAAUACGGAGGGUUUAAGGAGUGCCUUGGCCCGUUUACGCAACAGAACGGGCCCCUCAGUUCCA------CAAC---AACC------CACACCGCCAACCUCUCCCCUGCCUACCU ..........((((((((...((.(((....(((((((((........))))))))).....(((...------.)))---....------))).))...)))).))))........... ( -31.80) >consensus AAGACAAUACGGAGGGUUUACGGAGUGCUUUGGCCCGUUUACGCAACAGAACGGGCCCCUCAAUUCCA______CAAC___AACC______CACUCCGCCAGCCUCACCAUUGCCCA___ ..........((.(((((..(((.(((....(((((((((........)))))))))......((..........))..............))).)))..)))).).))........... (-23.69 = -23.47 + -0.22)

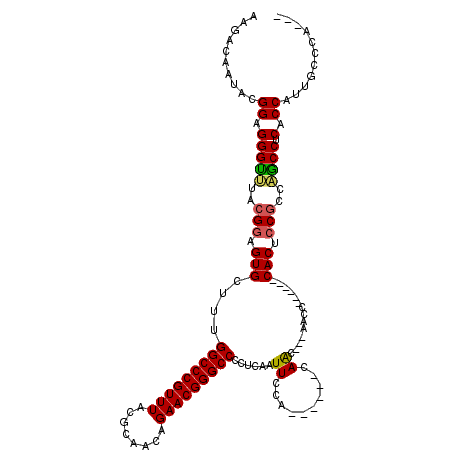
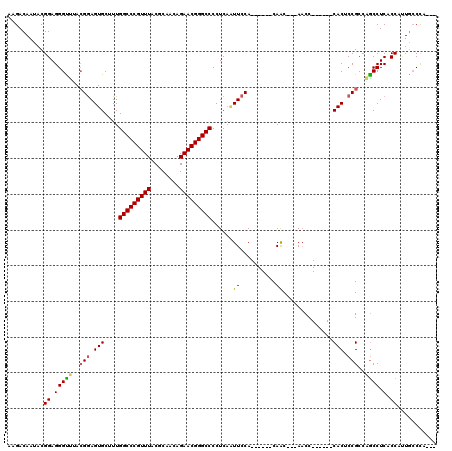
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:43 2006