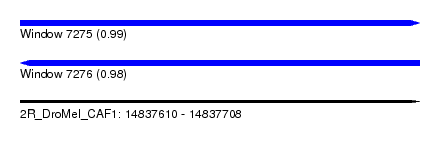
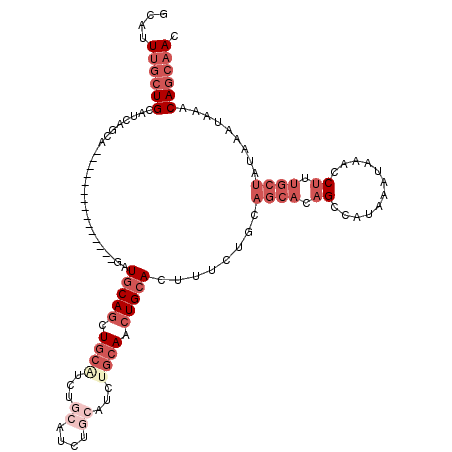
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,837,610 – 14,837,708 |
| Length | 98 |
| Max. P | 0.992808 |

| Location | 14,837,610 – 14,837,708 |
|---|---|
| Length | 98 |
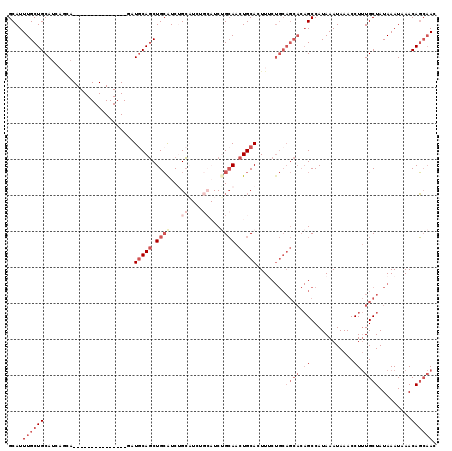
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -21.43 |
| Energy contribution | -24.16 |
| Covariance contribution | 2.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.17 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
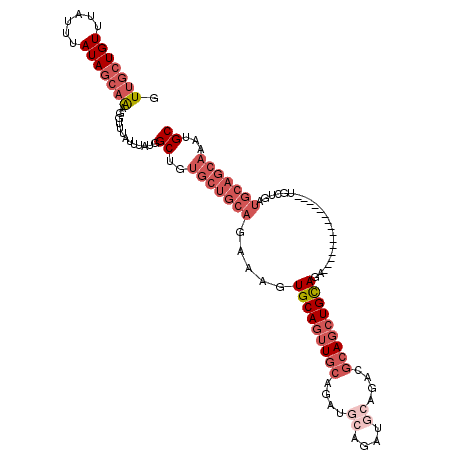
>2R_DroMel_CAF1 14837610 98 + 20766785 GUUGCUGUUUAUUUAUAGCAAAGGUUUAUUUAUGGCUGUGCUGCAGAAAGUGCAGUUGCCGA------UGCAGACGCAGCUGCAGA---------------UGCUGAUGCAGCAAAUGC .(((((((......)))))))............((((((((((((....(.((....)))..------))))).)))))))(((..---------------(((((...)))))..))) ( -35.70) >DroSec_CAF1 10028 104 + 1 GUUGCUGUUUAUUUAUAGCAAAGGUUUAUUUAUGGCUGUGCUGCAGAAAGUGCAGUUGCAGAUGCAGAUGCAGACGCAGCUGCAGA---------------UGCUGAUGCAGCAAAUGC .(((((((......))))))).............((..(((((((...(((((((((((...(((....)))...))))))))...---------------.)))..)))))))...)) ( -39.50) >DroSim_CAF1 10157 104 + 1 GUUGCUGUUUAUUUAUAGCAAAGGUUUAUUUAUGGCUGUGCUGCAGAAAGUGCAGUUGCAGAUGCAGAUGCAGACGCAGCUGCAGA---------------UGCUGAUGCAGCAAAUGC .(((((((......))))))).............((..(((((((...(((((((((((...(((....)))...))))))))...---------------.)))..)))))))...)) ( -39.50) >DroEre_CAF1 10091 98 + 1 GUUGCUGUUUAUUUAUAGCAAAGGUUUAUUUAUGGCUGUGCUGCAGAAAGUGCAGUUGCAGAUGCAGAGGCUGAUGCAGCUGCAUC---------------------UGCAGCAAAUGC .(((((((......))))))).............((..(((((((((...((((((((((...((....))...))))))))))))---------------------)))))))...)) ( -43.20) >DroYak_CAF1 10326 119 + 1 GUUGCUGUUUAUUUAUAGCAAAGGUUUAUUUAUGGCUGUGCUGCAGCAAGUGCAGUUGCAGAUGCUGAUGCUGAUGCAGCUGUAUCUGUAUCUGUAUCUGCUGUUUCUGCAGCAAAUGC .(((((((......))))))).............((((((..((((((..(((((.(((((((((...(((....)))...))))))))).)))))..))))))...))))))...... ( -49.70) >DroAna_CAF1 12883 85 + 1 GU--AUGUUUAUUUAUAGCAGAGGUUUAUUUAUGGC-----------AACUGCAGAUUCUGA------AUCAGAUUCAGAUGCAUC---------------UGUGGAUGCAGCAAAUGC ..--.(((.........((((..(((.......)))-----------..))))....(((((------((...)))))))((((((---------------....)))))))))..... ( -20.50) >consensus GUUGCUGUUUAUUUAUAGCAAAGGUUUAUUUAUGGCUGUGCUGCAGAAAGUGCAGUUGCAGAUGCAGAUGCAGACGCAGCUGCAGA_______________UGCUGAUGCAGCAAAUGC .(((((((......))))))).............((..(((((((.....(((((((((....((....))....))))))))).......................)))))))...)) (-21.43 = -24.16 + 2.72)

| Location | 14,837,610 – 14,837,708 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -11.33 |
| Energy contribution | -14.22 |
| Covariance contribution | 2.89 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.89 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
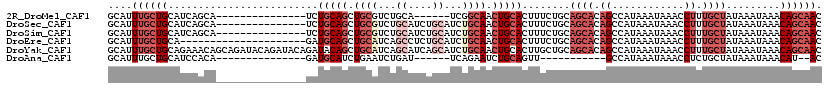
>2R_DroMel_CAF1 14837610 98 - 20766785 GCAUUUGCUGCAUCAGCA---------------UCUGCAGCUGCGUCUGCA------UCGGCAACUGCACUUUCUGCAGCACAGCCAUAAAUAAACCUUUGCUAUAAAUAAACAGCAAC (((..(((((...)))))---------------..))).((((.(.(((((------...((....))......)))))).)))).............(((((..........))))). ( -26.70) >DroSec_CAF1 10028 104 - 1 GCAUUUGCUGCAUCAGCA---------------UCUGCAGCUGCGUCUGCAUCUGCAUCUGCAACUGCACUUUCUGCAGCACAGCCAUAAAUAAACCUUUGCUAUAAAUAAACAGCAAC (((..(((((...)))))---------------..))).((((((..((((..(((....)))..)))).....))))))..................(((((..........))))). ( -28.50) >DroSim_CAF1 10157 104 - 1 GCAUUUGCUGCAUCAGCA---------------UCUGCAGCUGCGUCUGCAUCUGCAUCUGCAACUGCACUUUCUGCAGCACAGCCAUAAAUAAACCUUUGCUAUAAAUAAACAGCAAC (((..(((((...)))))---------------..))).((((((..((((..(((....)))..)))).....))))))..................(((((..........))))). ( -28.50) >DroEre_CAF1 10091 98 - 1 GCAUUUGCUGCA---------------------GAUGCAGCUGCAUCAGCCUCUGCAUCUGCAACUGCACUUUCUGCAGCACAGCCAUAAAUAAACCUUUGCUAUAAAUAAACAGCAAC ((...(((((((---------------------(((((((.((((...((....))...)))).)))))...)))))))))..)).............(((((..........))))). ( -31.30) >DroYak_CAF1 10326 119 - 1 GCAUUUGCUGCAGAAACAGCAGAUACAGAUACAGAUACAGCUGCAUCAGCAUCAGCAUCUGCAACUGCACUUGCUGCAGCACAGCCAUAAAUAAACCUUUGCUAUAAAUAAACAGCAAC ((...((((((((.....((((...(((((.(.(((...((((...))))))).).)))))...)))).....))))))))..)).............(((((..........))))). ( -32.40) >DroAna_CAF1 12883 85 - 1 GCAUUUGCUGCAUCCACA---------------GAUGCAUCUGAAUCUGAU------UCAGAAUCUGCAGUU-----------GCCAUAAAUAAACCUCUGCUAUAAAUAAACAU--AC (((.((((((((((....---------------))))))(((((((...))------)))))....)))).)-----------))..............................--.. ( -19.40) >consensus GCAUUUGCUGCAUCAGCA_______________GAUGCAGCUGCAUCUGCAUCUGCAUCUGCAACUGCACUUUCUGCAGCACAGCCAUAAAUAAACCUUUGCUAUAAAUAAACAGCAAC ....((((((.........................(((((.((((...((....))...)))).)))))........((((.((............)).)))).........)))))). (-11.33 = -14.22 + 2.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:33 2006