| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,834,809 – 14,834,916 |
| Length | 107 |
| Max. P | 0.727791 |

| Location | 14,834,809 – 14,834,916 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.14 |
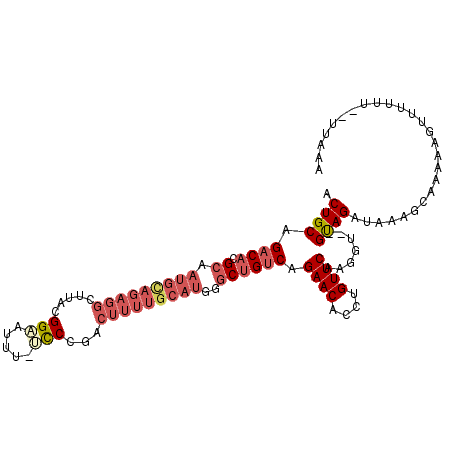
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -18.54 |
| Energy contribution | -20.27 |
| Covariance contribution | 1.72 |
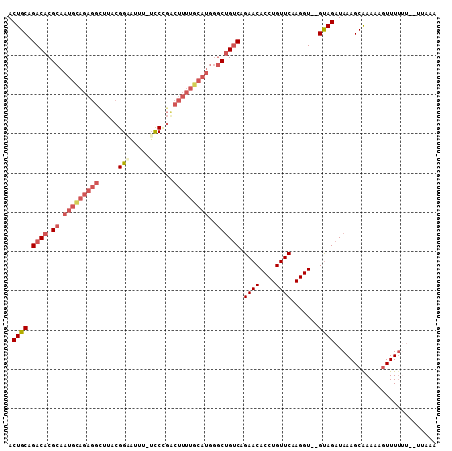
| Combinations/Pair | 1.15 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727791 |
| Prediction | RNA |
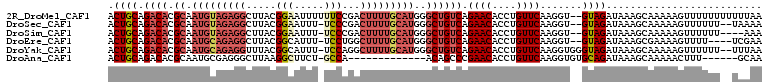
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14834809 107 - 20766785 ACUGCAGACACGCAAUGUAGAGGCUUACGGAAUUUUUUCCGACUUUUGCAUGGGCUGUCAGAACACCUGUUCAAGGU--GUAGAUAAAGCAAAAAGUUUUUUUUUUUAA ..(((.((((.((.(((((((((....(((((....))))).)))))))))..))))))...((((((.....))))--)).......))).................. ( -33.00) >DroSec_CAF1 7248 104 - 1 ACUGCAGACACGCAAUGUAGAGGCUUACGGAAUUU-UCCCGACUUUUGCAUGGGCUGUCAGAACACCUGUUCAAGGU--GUAGAUAAAGCAAAAAGUUUUUU--UAAAA ..(((.((((.((.(((((((((....(((.....-..))).)))))))))..))))))...((((((.....))))--)).......)))...........--..... ( -29.80) >DroSim_CAF1 7356 102 - 1 ACUGCAGACACGCAAUGUAGAGGCUUACGGAAUUU-UCCCGACUUUUGCAUGGGCUGUCAGAACACCUGUUCAAGGU--GUAGAUAAAGCAAAAAGUUUUUU----AAA ..(((.((((.((.(((((((((....(((.....-..))).)))))))))..))))))...((((((.....))))--)).......)))...........----... ( -29.80) >DroEre_CAF1 7309 102 - 1 ACUGCAGACACGCAAUGCAGAGGCUUACGGCAUUU-UCCUGGCUUUUGCAUGGGCUGUCAGAACACCUGUUCAAGGU--GUAGAUAAAGCGAAAAGUUUU----UCGAA ...((.((((.((.(((((((((((...((.....-.)).)))))))))))..))))))...((((((.....))))--)).......))((((....))----))... ( -35.10) >DroYak_CAF1 7480 106 - 1 ACUGCAGACACGCAAUGCAGAGGUUUACGGCAUUU-UCCAGGCUUUUGCAUGGGCUGUCAGAACACCUGUUCAAGGUGGGUAGAUAAAGCAAAAAGUUUUUU--UUUAA .((((.((((.((.((((((((((((..((.....-.))))))))))))))..))))))....(((((.....))))).))))..(((((.....)))))..--..... ( -33.30) >DroAna_CAF1 10291 89 - 1 ACUGCAGACACGCAAUGCGAGGGCUUAAGGCUUCU-GCCA-------------ACAGCCCGAACACCUGUUCAAGGUGUGCAGAUAAAGCAAAAACUUU------GCAA .(((((.((.(((...))).(((((...(((....-))).-------------..)))))((((....))))...)).))))).....((((.....))------)).. ( -33.40) >consensus ACUGCAGACACGCAAUGCAGAGGCUUACGGAAUUU_UCCCGACUUUUGCAUGGGCUGUCAGAACACCUGUUCAAGGU__GUAGAUAAAGCAAAAAGUUUUUU__UUAAA .((((.((((.((.(((((((((.....(((.....)))...)))))))))..)))))).((((....)))).......)))).......................... (-18.54 = -20.27 + 1.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:31 2006