| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,834,505 – 14,834,619 |
| Length | 114 |
| Max. P | 0.579490 |

| Location | 14,834,505 – 14,834,619 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.11 |
| Mean single sequence MFE | -13.50 |
| Consensus MFE | -10.26 |
| Energy contribution | -10.28 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
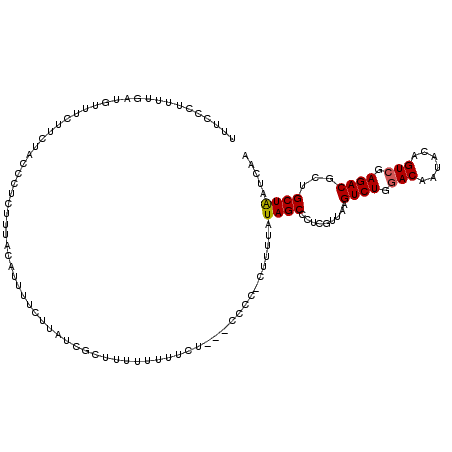
>2R_DroMel_CAF1 14834505 114 - 20766785 UUUCCAUUUUGAUGUUUCUUCUACCCUCUUUACAUUUUCUUAUCGCUUUUUUUCCC---CCCUCUUUUUAUAGCCCUCGUUAAGUCUGGACAAUACAGUCGAGACGCUGCUAAUCAA ........(((((...............................(((.........---............)))....((...((((.(((......))).))))...))..))))) ( -13.70) >DroSec_CAF1 6946 112 - 1 UUUCCCUUUUGAUGUUUCUUCUACGCUCUUUACAUUUUCUUAUCGCUUUU-UUUCU---CCCC-CUUUUAUAGCCCUCGUUAAGUCUGGACAAUACAGUCGAGACGCUGCUAAUCAA ........(((((...............................(((...-.....---....-.......)))....((...((((.(((......))).))))...))..))))) ( -13.81) >DroSim_CAF1 7053 113 - 1 UUUCCUUUUUGAUGUUUCUUCUACGCUCUUUACAUUUUCUUAUCGCUUUUUUUUCU---CCCC-CUUUUAUAGCCCUCGUUAAGUCUGGACAAUACAGUCGAGACGCUGCUAAUCAA ........(((((...............................(((.........---....-.......)))....((...((((.(((......))).))))...))..))))) ( -13.75) >DroEre_CAF1 7008 112 - 1 UUUUUCUCUUGUUGUUUCCCUUACCCUCUUUACAUUUUCUCAUAGUUCUUUCU--C--UUCUC-CUUUUAUAGCCCUCGUUAAGUCUGGACAAUACAGUCGAGACGCUGCUAAUCAA ..........................................((((.......--.--.....-......((((....)))).((((.(((......))).))))...))))..... ( -12.30) >DroYak_CAF1 7173 114 - 1 UUUUCUUUUAGAUGUUUCCUUUACCCUCUUUACAUUUUCUCAUCGCUUUUUUUUCU--UUCUA-CUUUUAUAGCCCUCGUUAAGUCUGGACAAUACAGUCGAGACGCUGCUAAUCAA ..........((((..........................))))((..........--.....-......((((....)))).((((.(((......))).)))))).......... ( -13.87) >DroAna_CAF1 10034 91 - 1 --CU-------------------CCUUUCUUACGUUCUCUAU----UUUUUUUUCAUAAUUUC-UUUAAAUAGCCAUCUUUAAGUCUGGACAAUACGGUGGAGACGCUGCUGAUCAA --.(-------------------((...((((......((((----((...............-...)))))).......))))...))).....(((((....)))))........ ( -13.59) >consensus UUUCCCUUUUGAUGUUUCUUCUACCCUCUUUACAUUUUCUUAUCGCUUUUUUUUCU___CCCC_CUUUUAUAGCCCUCGUUAAGUCUGGACAAUACAGUCGAGACGCUGCUAAUCAA ......................................................................((((.........((((.(((......))).))))...))))..... (-10.26 = -10.28 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:30 2006