| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,833,563 – 14,833,655 |
| Length | 92 |
| Max. P | 0.879322 |

| Location | 14,833,563 – 14,833,655 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.99 |
| Mean single sequence MFE | -34.23 |
| Consensus MFE | -16.98 |
| Energy contribution | -17.23 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
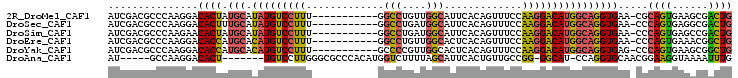
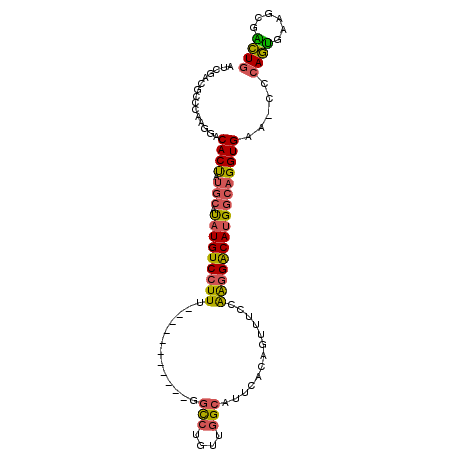
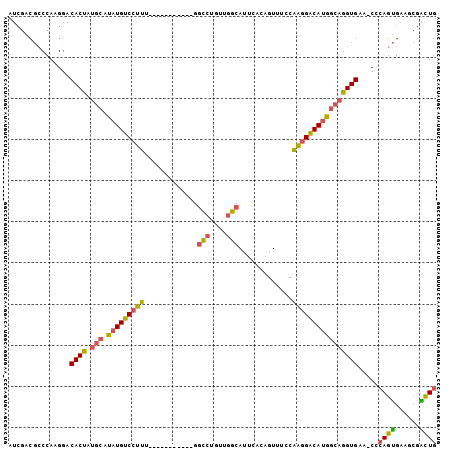
>2R_DroMel_CAF1 14833563 92 + 20766785 CAGUCGCUUCACUGCG-UUCACCUGCCAUGUCCUUGGAAACUGUGAAUGCCAACAGGCC-----------AAAGGACAUAUGCAUAGUGUCCUUGGGCGUCGAU ..(((((......(((-(((((...(((......))).....))))))))......(((-----------.((((((((.......)))))))).)))).)))) ( -32.30) >DroSec_CAF1 6031 92 + 1 CAGUCGCCUCACUGGG-UUCACCUGCCAUGUCCUUGGAAACUGUGAAUGCCAUCAGGCC-----------AAAGGACAUAUGCAAAGUGUCCUUGGGCGUCGAU ..(.(((((....(((-..(((.(((.((((((((((...(((((.....)).))).))-----------.))))))))..)))..))).))).))))).)... ( -33.50) >DroSim_CAF1 6126 92 + 1 CAGUCGGCUCACUGGG-UUCACCUGCCAUGUCCUUGGAAACUGUGAAUGCCAUCAGGCC-----------AAAGGACAUAUGCAUAGUGUUCUUGGGCGUCGAU ..((((((.(.(..((-..(((.(((.((((((((((...(((((.....)).))).))-----------.))))))))..)))..)))..))..)).)))))) ( -34.90) >DroEre_CAF1 6043 92 + 1 CAGCCGUUUCACUGGG-UUCACCUGCCAUGUCCUUGGAAACUGUGAGUGCCAACAGGCC-----------AAAGGACAUGUGCAUGGUGUCCUUGGGCGUCGAU ....((.....(..((-..((((((((((((((((((...((((........)))).))-----------.))))))))).))).))))..))..)....)).. ( -39.40) >DroYak_CAF1 6222 92 + 1 CAGCCGCUUCACUGGG-CUCACCUGCCAUGUCCUUGGAAACUGUGAGUGCCAACGGGGC-----------AAAGGACAUGUGCAUGGUGUCCUUGGGCGUCGAU ..(.(((((....(((-..(((((((((((((((((....)......((((.....)))-----------)))))))))).))).)))).))).))))).)... ( -39.10) >DroAna_CAF1 9063 90 + 1 CAAAUUUUACCUUCCGUUGCACCUGG-AUGCC-CCGGCAACAGUGAAUGCUAAAAGACCAUGUGGGCGCCCAAGGACA-------AGUGUCCUUGGC-----AU ..............((((.(((.(((-.(((.-...)))..(((....)))......))).))))))).((((((((.-------...)))))))).-----.. ( -26.20) >consensus CAGUCGCUUCACUGGG_UUCACCUGCCAUGUCCUUGGAAACUGUGAAUGCCAACAGGCC___________AAAGGACAUAUGCAUAGUGUCCUUGGGCGUCGAU ..(((......(((...(((((...(((......))).....)))))......)))...............((((((((.......)))))))).)))...... (-16.98 = -17.23 + 0.25)

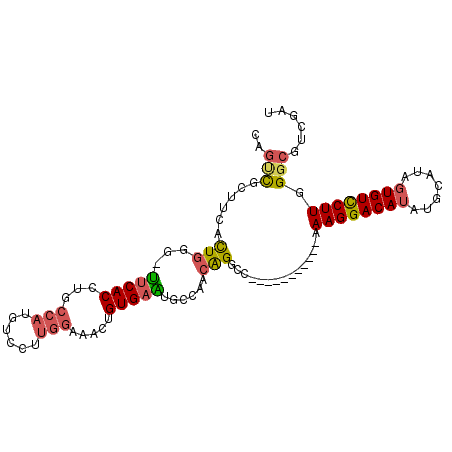
| Location | 14,833,563 – 14,833,655 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.99 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.34 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14833563 92 - 20766785 AUCGACGCCCAAGGACACUAUGCAUAUGUCCUUU-----------GGCCUGUUGGCAUUCACAGUUUCCAAGGACAUGGCAGGUGAA-CGCAGUGAAGCGACUG .(((.(((....(..((((.(((.(((((((((.-----------((.((((........))))...))))))))))))))))))..-)...)))...)))... ( -28.20) >DroSec_CAF1 6031 92 - 1 AUCGACGCCCAAGGACACUUUGCAUAUGUCCUUU-----------GGCCUGAUGGCAUUCACAGUUUCCAAGGACAUGGCAGGUGAA-CCCAGUGAGGCGACUG .....((((((.((.(((((.((.(((((((((.-----------((.(((.((.....)))))...))))))))))))))))))..-))...)).)))).... ( -32.20) >DroSim_CAF1 6126 92 - 1 AUCGACGCCCAAGAACACUAUGCAUAUGUCCUUU-----------GGCCUGAUGGCAUUCACAGUUUCCAAGGACAUGGCAGGUGAA-CCCAGUGAGCCGACUG .(((..((.((....((((.(((.(((((((((.-----------((.(((.((.....)))))...))))))))))))))))))..-.....)).)))))... ( -25.10) >DroEre_CAF1 6043 92 - 1 AUCGACGCCCAAGGACACCAUGCACAUGUCCUUU-----------GGCCUGUUGGCACUCACAGUUUCCAAGGACAUGGCAGGUGAA-CCCAGUGAAACGGCUG ......(((...((.((((.(((.(((((((((.-----------((.((((........))))...))))))))))))))))))..-.)).(.....)))).. ( -32.30) >DroYak_CAF1 6222 92 - 1 AUCGACGCCCAAGGACACCAUGCACAUGUCCUUU-----------GCCCCGUUGGCACUCACAGUUUCCAAGGACAUGGCAGGUGAG-CCCAGUGAAGCGGCUG ......(((......((((.(((.((((((((((-----------(((.....))))............)))))))))))))))).(-(........))))).. ( -32.00) >DroAna_CAF1 9063 90 - 1 AU-----GCCAAGGACACU-------UGUCCUUGGGCGCCCACAUGGUCUUUUAGCAUUCACUGUUGCCGG-GGCAU-CCAGGUGCAACGGAAGGUAAAAUUUG ..-----.((((((((...-------.))))))))(((((...(((.(((..(((((.....)))))..))-).)))-...)))))..(....).......... ( -31.20) >consensus AUCGACGCCCAAGGACACUAUGCAUAUGUCCUUU___________GGCCUGUUGGCAUUCACAGUUUCCAAGGACAUGGCAGGUGAA_CCCAGUGAAGCGACUG ...............((((.(((.(((((((((.............(((....))).............)))))))))))))))).....((((......)))) (-21.62 = -21.34 + -0.29)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:29 2006