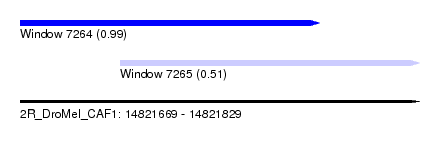
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,821,669 – 14,821,829 |
| Length | 160 |
| Max. P | 0.990662 |

| Location | 14,821,669 – 14,821,789 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -27.24 |
| Energy contribution | -26.88 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14821669 120 + 20766785 AAACAUAGUUAACGAACAAAUUUUGCUAACACGACUGGCAAAUGCAGCCAUUGAUAUUUAUGCAAUGGUUGUGACGCAAUCGCGAUCAUCCCGUGCAGUCAAUCUGAAUCUGCCAACUGC .......((((.((((.....)))).)))).....(((((.((.(((..((((((...((((..((((((((((.....))))))))))..))))..))))))))).)).)))))..... ( -30.10) >DroSec_CAF1 23598 120 + 1 AAACAUAGUUAACGAACAAAUUUUGCUAACACGACUGGCAAAUGCAGCCAUUGAUAUUUAUGCAAUGGUUGUGACGCAAUCACGAUCAUCCCGUGCAGUCACUCUGAAUCUGCCAACUGC .......((((.((((.....)))).)))).....(((((...((((((((((.........))))))))))(((.....((((.......))))..)))..........)))))..... ( -29.00) >DroSim_CAF1 17808 120 + 1 AAACAUAGUUGACGAACAAAUUUUGCUAACACGACUGGCAAAUGCAGCCAUUGAUAUUUAUGCAAUGGUUGUGACGCAAUCACGAUCAUCCCGUGCAGUCACUCUGAAUCUGCCAACUGC .....((((((..........(((((((.......))))))).((((((((((.........))))))..(((((.....((((.......))))..))))).......)))))))))). ( -30.00) >DroEre_CAF1 16458 120 + 1 AAAUAUAGUUAACGAACAAAUUUUGCUAACACGACUGGCCAAUGCAGCCAUUGAUAUAUACGCGAUGGUUGUGACGCAAUCGCGAUCAUCUCGUGCAGUCAGUCUAAAUCUGCCAACUGC .......((((.((((.....)))).))))..((((((((((((....))))).....((((.(((((((((((.....))))))))))).))))..)))))))................ ( -34.70) >DroYak_CAF1 17643 120 + 1 AAAUAUAGUUAACGAACAAAUUUUGCUAACACGACUGGCCAAUGCAGCCAUUGAUAUAUACGCGAUGGUUGUGACGCAAUCCCGAUCAUCUCGUGCAGUCAGUCUCAAUCUGCCAACUGC .......((((.((((.....)))).))))..((((((((((((....))))).....((((.((((((((.((.....)).)))))))).))))..)))))))................ ( -30.30) >consensus AAACAUAGUUAACGAACAAAUUUUGCUAACACGACUGGCAAAUGCAGCCAUUGAUAUUUAUGCAAUGGUUGUGACGCAAUCACGAUCAUCCCGUGCAGUCACUCUGAAUCUGCCAACUGC .......((((.((((.....)))).)))).....((((....((((((((((.........))))))))))(((.....((((.......))))..)))...........))))..... (-27.24 = -26.88 + -0.36)

| Location | 14,821,709 – 14,821,829 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -25.41 |
| Energy contribution | -25.45 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14821709 120 + 20766785 AAUGCAGCCAUUGAUAUUUAUGCAAUGGUUGUGACGCAAUCGCGAUCAUCCCGUGCAGUCAAUCUGAAUCUGCCAACUGCACAACACGAGCUCAACAUGACCAAGGCACUCACUAUUCAG ...((((((((((.........))))))))))(((((....))).)).....((((.((((...(((.(((((.....)))......))..)))...))))....))))........... ( -27.60) >DroSec_CAF1 23638 120 + 1 AAUGCAGCCAUUGAUAUUUAUGCAAUGGUUGUGACGCAAUCACGAUCAUCCCGUGCAGUCACUCUGAAUCUGCCAACUGCACAACACGAGCUCAACAUGACCAAGGCACUCACUAUUCAG ......(((.(((....(((((..((((((((((.....))))))))))...(((((((................))))))).............))))).))))))............. ( -26.89) >DroSim_CAF1 17848 120 + 1 AAUGCAGCCAUUGAUAUUUAUGCAAUGGUUGUGACGCAAUCACGAUCAUCCCGUGCAGUCACUCUGAAUCUGCCAACUGCACAACACGAGCUCAACAUGACCAAGGCACUCACUAUUCAG ......(((.(((....(((((..((((((((((.....))))))))))...(((((((................))))))).............))))).))))))............. ( -26.89) >DroEre_CAF1 16498 120 + 1 AAUGCAGCCAUUGAUAUAUACGCGAUGGUUGUGACGCAAUCGCGAUCAUCUCGUGCAGUCAGUCUAAAUCUGCCAACUGCACAACACGAGCUCAACAUGACCAAGGCGCUCACUAUUCAG ...((.(((.(((........(((((((((((((.....)))))))))))..((((((((((.......)))...))))))).......))((.....)).))))))))........... ( -34.60) >DroYak_CAF1 17683 120 + 1 AAUGCAGCCAUUGAUAUAUACGCGAUGGUUGUGACGCAAUCCCGAUCAUCUCGUGCAGUCAGUCUCAAUCUGCCAACUGCACAACACGAGCUCAACAUGACCAAGGCCCUCACAAUUCAG ......(((.(((........((((((((((.((.....)).))))))))..((((((((((.......)))...))))))).......))((.....)).))))))............. ( -28.00) >consensus AAUGCAGCCAUUGAUAUUUAUGCAAUGGUUGUGACGCAAUCACGAUCAUCCCGUGCAGUCACUCUGAAUCUGCCAACUGCACAACACGAGCUCAACAUGACCAAGGCACUCACUAUUCAG ......(((.(((........((.((((((((((.....))))))))))...(((((((................))))))).......))((.....)).))))))............. (-25.41 = -25.45 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:23 2006