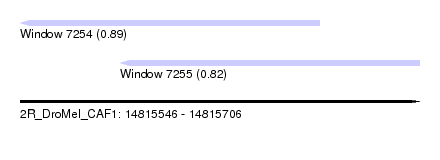
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,815,546 – 14,815,706 |
| Length | 160 |
| Max. P | 0.891185 |

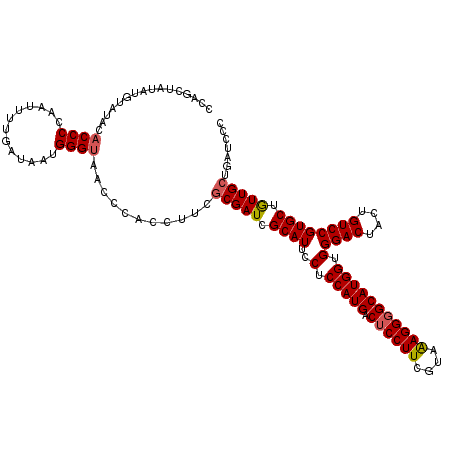
| Location | 14,815,546 – 14,815,666 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -39.48 |
| Consensus MFE | -36.84 |
| Energy contribution | -36.80 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.891185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14815546 120 - 20766785 CCACCUUCGCGAUCGCAUUCCUCCAUGACACCUUCGUAAAGGGGCAUGGUGGGACUACUGUCCGUGCUAUUGCUGAUCCCAAUGUUGUUAUUGUCGCUGACGGCAUGGUUGUUACUAUAG ........(((((.((((..(.(((((...((((....))))..))))).)((((....)))))))).))))).....(((.((((((((.......)))))))))))............ ( -36.10) >DroSec_CAF1 17393 120 - 1 CUACCUUCGCGAUCGCAUUCCUCCAUGACUCCUUCGUAAAGGGGCAUGGUGGGACUACUGUCCGUGCUGUUGCCGAUCCCAAUGUUGUUAUUGUCGCUGACGGCAUGGUUGUUACUAUAG ........((....)).((((.(((((.((((((....))))))))))).))))..((...((((((((((..(((...(((((....))))))))..))))))))))..))........ ( -41.30) >DroSim_CAF1 11666 120 - 1 CCACCUUCGCGAUCGCAUUCCUCCAUGACUCCUUCGUAAAGGGGCAUGGUGGGACUACUGUCCGUGCUGUUGCUGAUCCCAAUGUUGUUAUUGUCGCUGACGGCAUGGUUGUUACUAUAG ........((....)).((((.(((((.((((((....))))))))))).))))..((...((((((((((((.((...(((((....))))))))).))))))))))..))........ ( -39.80) >DroEre_CAF1 10270 120 - 1 GCACCUUCGCGAUCGCAUUCCUCCAUGACUCCUUCGUAAAGGGGCAUGGUGGGACUACUGUCCGUGCUGUUGCUGAUCCCAAUGUUGUUAUUGUCGCUGACGGCAUGGUUGUUACUAUAG (((((...(((((.((((..(.(((((.((((((....))))))))))).)((((....)))))))).)))))........(((((((((.......))))))))))).)))........ ( -40.20) >DroYak_CAF1 11543 120 - 1 CCACCUUCUCGAUCGCAUUCCUCCAUGACUCCUUCGUAGAGGGGCAUGGUGGGACUACUGUCCGUGCUGUUGCUGAUCCCAAUGUUGUUGUUGUCGCUGACGGCAUGAUUGUUACUAUAG .........(((((......(.(((((.((((((....))))))))))).)((((....))))((((((((((.(((..((((...))))..))))).)))))))))))))......... ( -40.00) >consensus CCACCUUCGCGAUCGCAUUCCUCCAUGACUCCUUCGUAAAGGGGCAUGGUGGGACUACUGUCCGUGCUGUUGCUGAUCCCAAUGUUGUUAUUGUCGCUGACGGCAUGGUUGUUACUAUAG ........((((((......(.(((((.((((((....))))))))))).)((((....))))((((((((..(((...(((((....))))))))..))))))))))))))........ (-36.84 = -36.80 + -0.04)

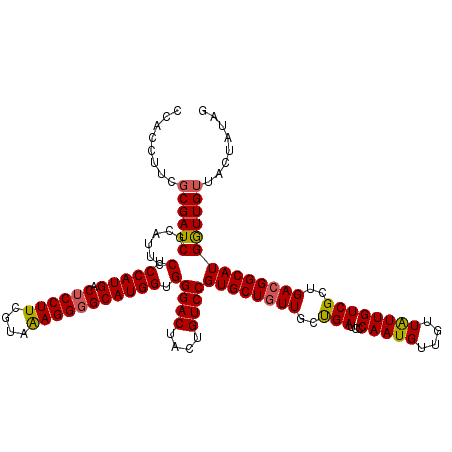
| Location | 14,815,586 – 14,815,706 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -31.46 |
| Energy contribution | -31.74 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14815586 120 - 20766785 CCAGCUAUAUGUAUGCACCCCAAUUUUGAUAAUGGGUAACCCACCUUCGCGAUCGCAUUCCUCCAUGACACCUUCGUAAAGGGGCAUGGUGGGACUACUGUCCGUGCUAUUGCUGAUCCC .((((.(((.(((((.((((((..........))))...((((((...((....)).(((((..((((.....))))..)))))...))))))......)).)))))))).))))..... ( -35.00) >DroSec_CAF1 17433 120 - 1 CCAGCUAUAUGUGUACACCCCAAUUUUCAUAAUGGGUAACCUACCUUCGCGAUCGCAUUCCUCCAUGACUCCUUCGUAAAGGGGCAUGGUGGGACUACUGUCCGUGCUGUUGCCGAUCCC ..................((((..........))))..........(((((((.((((..(.(((((.((((((....))))))))))).)((((....)))))))).)))).))).... ( -33.90) >DroSim_CAF1 11706 120 - 1 CCAGCUAUAUGUAUGCACCCCAAUUUUGAUAAUGGGUAACCCACCUUCGCGAUCGCAUUCCUCCAUGACUCCUUCGUAAAGGGGCAUGGUGGGACUACUGUCCGUGCUGUUGCUGAUCCC .((((.(((.(((((.((((((..........))))...((((((...((....))........(((.((((((....)))))))))))))))......)).)))))))).))))..... ( -37.60) >DroEre_CAF1 10310 120 - 1 CCUGCUAUAUGUUUACUCCCUCAUUCUGAUAAAGGGUAACGCACCUUCGCGAUCGCAUUCCUCCAUGACUCCUUCGUAAAGGGGCAUGGUGGGACUACUGUCCGUGCUGUUGCUGAUCCC .........(((((((((..((.....))....)))))).))).....(((((.((((..(.(((((.((((((....))))))))))).)((((....)))))))).)))))....... ( -35.30) >DroYak_CAF1 11583 120 - 1 CCUGCUAUAUGUGGACACCCCCAUUCUGAUAAAGGGUAACCCACCUUCUCGAUCGCAUUCCUCCAUGACUCCUUCGUAGAGGGGCAUGGUGGGACUACUGUCCGUGCUGUUGCUGAUCCC ..........((((..((((.............))))...))))......(((((((...(.(((((.((((((....))))))))))).)((((....)))).......))).)))).. ( -36.22) >consensus CCAGCUAUAUGUAUACACCCCAAUUUUGAUAAUGGGUAACCCACCUUCGCGAUCGCAUUCCUCCAUGACUCCUUCGUAAAGGGGCAUGGUGGGACUACUGUCCGUGCUGUUGCUGAUCCC ................((((.............))))...........(((((.((((..(.(((((.((((((....))))))))))).)((((....)))))))).)))))....... (-31.46 = -31.74 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:12 2006