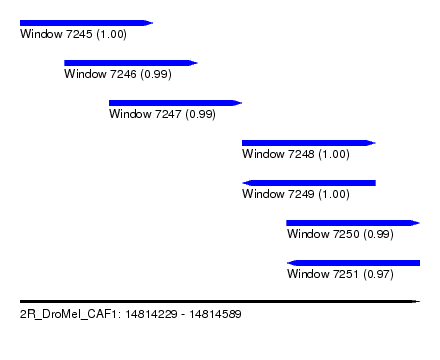
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,814,229 – 14,814,589 |
| Length | 360 |
| Max. P | 0.999788 |

| Location | 14,814,229 – 14,814,349 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -48.02 |
| Consensus MFE | -46.34 |
| Energy contribution | -46.18 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14814229 120 + 20766785 UCCCGCCAGCCGCAGCGGGAGCUAUGAGCAGCUGGAGCUGCUGGAUCACGCAACGCAUCCACAGCAACAACAGGAAGCCAGCAGUGGCAAGGCCAAGCCGAGCUGCCAUUGCACCAACAU ((((((........))))))((.(((.((((((((.((((((((.....((...)).(((............)))..))))))))(((...)))...)).))))))))).))........ ( -47.10) >DroSec_CAF1 16076 120 + 1 UCCCGCCAGCCGCAGCGGGAGCUAUGAGCAGCUGGAGCUGUUGGAUCACGCAACGCAUCCACAGCAGCAACAGGAGGCCACCAGUGGCAAGGCCAAGCCUAGCUGUCAUUGCACCAACAU ((((((........))))))((.((((.(((((((.(((((((((((.......).)))))..)))))....((..((((....))))....))....))))))))))).))........ ( -48.70) >DroSim_CAF1 10349 120 + 1 UCCCGCCAGCCGCAGCGGGAGCUAUGAGCAGCUGGAGCUGCUGGAUCACGCAACGCAUCCACAGCAGCAACAGGAGGCCACCAGUGGCAAGGCCAAGCCUAGCUGUCAUUGCACCAACAU ((((((........))))))((.((((.(((((((.(((((((......((...)).....)))))))....((..((((....))))....))....))))))))))).))........ ( -50.20) >DroEre_CAF1 8953 120 + 1 UCCCGCCAGCCGCAGCGGGAGCUAUGAGCAGCUGGAGCUGCUGGAUCACGCAACGCAUCCACAGCAGGAACAGGAAGCCACCAGUGGCAAGGCCAAGCCGAGCUGUCAUUGCACCAACAU ((((((........))))))((.((((.(((((((.((((.((((((.......).))))))))).......((..((((....))))....))...)).))))))))).))........ ( -45.70) >DroYak_CAF1 10223 120 + 1 UCCCGCUAGCCGCAGCGGGAGCUAUGAGCAGCUGGAGCUGCUGGAUCACGCAACGCAUCCACAGCAGGAACAGGAGGCCACCAGUGGCAAGGCCAAGCCGAGCUGUCAUUGCACCAACAU (((((((......)))))))((.((((.(((((((.((((.((((((.......).))))))))).......((..((((....))))....))...)).))))))))).))........ ( -48.40) >consensus UCCCGCCAGCCGCAGCGGGAGCUAUGAGCAGCUGGAGCUGCUGGAUCACGCAACGCAUCCACAGCAGCAACAGGAGGCCACCAGUGGCAAGGCCAAGCCGAGCUGUCAUUGCACCAACAU ((((((........))))))((.(((.((((((((.((((.((((((.......).))))))))).......((..((((....))))....))...)).))))))))).))........ (-46.34 = -46.18 + -0.16)

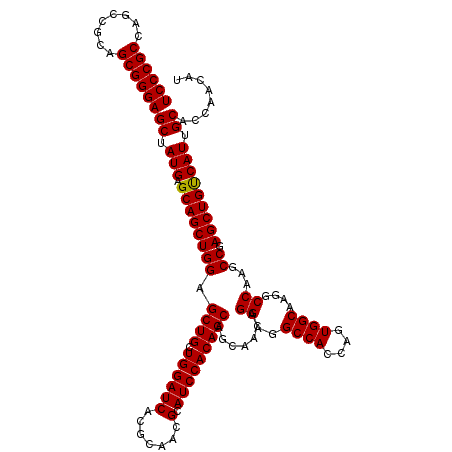
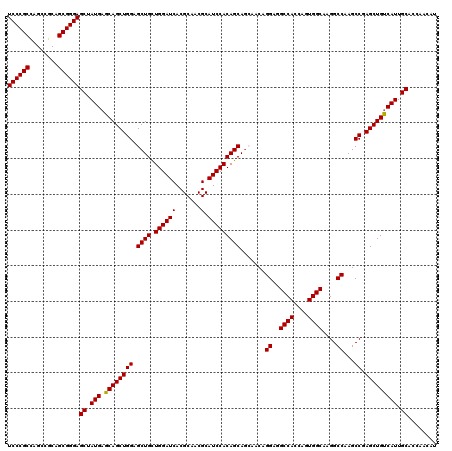
| Location | 14,814,269 – 14,814,389 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -34.98 |
| Energy contribution | -34.94 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14814269 120 + 20766785 CUGGAUCACGCAACGCAUCCACAGCAACAACAGGAAGCCAGCAGUGGCAAGGCCAAGCCGAGCUGCCAUUGCACCAACAUUAGUAUUAUGCACCUGUUUCAUGAGAUGAAGCAGGAGUUC .........(((..((.(((............))).))..(((((((((.(((...)))....)))))))))................))).((((((((((...))))))))))..... ( -38.30) >DroSec_CAF1 16116 120 + 1 UUGGAUCACGCAACGCAUCCACAGCAGCAACAGGAGGCCACCAGUGGCAAGGCCAAGCCUAGCUGUCAUUGCACCAACAUUAGUAUUAUGCACCUGUUUCAUGAGAUGAAACAGGAGUUC .........(((..(((...(((((...........((((....)))).((((...)))).)))))...)))....((....))....))).((((((((((...))))))))))..... ( -36.50) >DroSim_CAF1 10389 120 + 1 CUGGAUCACGCAACGCAUCCACAGCAGCAACAGGAGGCCACCAGUGGCAAGGCCAAGCCUAGCUGUCAUUGCACCAACAUUAGUAUUAUGCACCUGUUUCAUGAGAUGAAACAGGAGUUC .........(((..(((...(((((...........((((....)))).((((...)))).)))))...)))....((....))....))).((((((((((...))))))))))..... ( -36.50) >DroEre_CAF1 8993 120 + 1 CUGGAUCACGCAACGCAUCCACAGCAGGAACAGGAAGCCACCAGUGGCAAGGCCAAGCCGAGCUGUCAUUGCACCAACAUUAGUAUUAUGCACCUGUUUCAUGAGAUGAAGCAGGAGUUC .........(((..(((...(((((.((....((..((((....))))....))...))..)))))...)))....((....))....))).((((((((((...))))))))))..... ( -36.00) >DroYak_CAF1 10263 120 + 1 CUGGAUCACGCAACGCAUCCACAGCAGGAACAGGAGGCCACCAGUGGCAAGGCCAAGCCGAGCUGUCAUUGCACCAACAUUAGUAUUAUGCACCUGUUUCAUGAGAUGAAGCAGGAGUUC .........(((..(((...(((((.((....((..((((....))))....))...))..)))))...)))....((....))....))).((((((((((...))))))))))..... ( -37.10) >consensus CUGGAUCACGCAACGCAUCCACAGCAGCAACAGGAGGCCACCAGUGGCAAGGCCAAGCCGAGCUGUCAUUGCACCAACAUUAGUAUUAUGCACCUGUUUCAUGAGAUGAAGCAGGAGUUC .........(((..(((...(((((...........((((....))))..(((...)))..)))))...)))....((....))....))).((((((((((...))))))))))..... (-34.98 = -34.94 + -0.04)

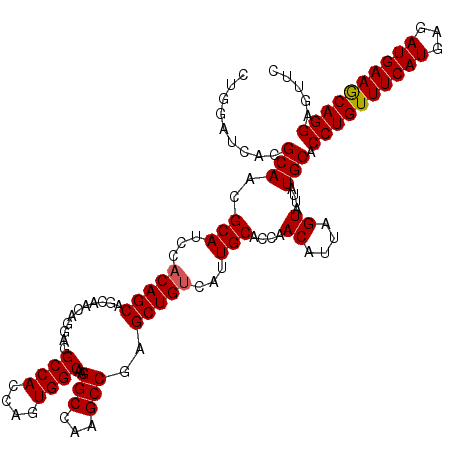
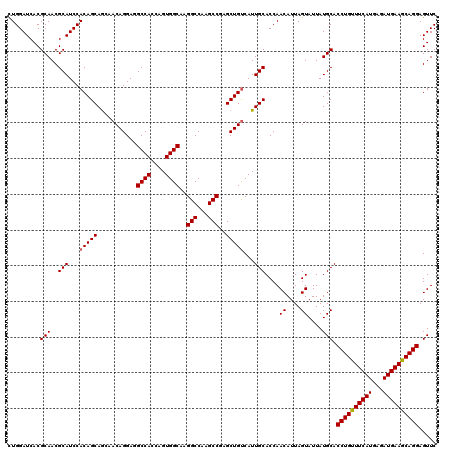
| Location | 14,814,309 – 14,814,429 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -39.62 |
| Consensus MFE | -34.96 |
| Energy contribution | -35.32 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14814309 120 + 20766785 GCAGUGGCAAGGCCAAGCCGAGCUGCCAUUGCACCAACAUUAGUAUUAUGCACCUGUUUCAUGAGAUGAAGCAGGAGUUCCCCACGAUACCCGAUGCCAUAGUGACGCAAUGCGUCAAUG (((((((((.(((...)))....))))))))).....((((.((((..(((.((((((((((...)))))))))).).....))..))))..))))......((((((...))))))... ( -41.90) >DroSec_CAF1 16156 120 + 1 CCAGUGGCAAGGCCAAGCCUAGCUGUCAUUGCACCAACAUUAGUAUUAUGCACCUGUUUCAUGAGAUGAAACAGGAGUUCCCCACGAUACCGGAUGCCAUUGUGACGCAAUGCGUCAAUG .((((((((((((...)))).(.((((..((((...((....))....))))((((((((((...))))))))))..........)))).)...))))))))((((((...))))))... ( -40.10) >DroSim_CAF1 10429 120 + 1 CCAGUGGCAAGGCCAAGCCUAGCUGUCAUUGCACCAACAUUAGUAUUAUGCACCUGUUUCAUGAGAUGAAACAGGAGUUCCCCACGAUACCCGACGCCAUUGUGACGCAAUGCGUCAAUG .(((((((.((((...))))....(((..((((...((....))....))))((((((((((...)))))))))).................))))))))))((((((...))))))... ( -40.10) >DroEre_CAF1 9033 120 + 1 CCAGUGGCAAGGCCAAGCCGAGCUGUCAUUGCACCAACAUUAGUAUUAUGCACCUGUUUCAUGAGAUGAAGCAGGAGUUCCCCACGAUACCCGAUGCCAUAGUGACGCAAUGCGUCAAUG ...((((((.(((...)))(((((.....((((...((....))....))))((((((((((...)))))))))))))))..............))))))..((((((...))))))... ( -38.00) >DroYak_CAF1 10303 120 + 1 CCAGUGGCAAGGCCAAGCCGAGCUGUCAUUGCACCAACAUUAGUAUUAUGCACCUGUUUCAUGAGAUGAAGCAGGAGUUCCCCACGAUACCCGAUGCCAUAGUGACGCAAUGCGUCAAUG ...((((((.(((...)))(((((.....((((...((....))....))))((((((((((...)))))))))))))))..............))))))..((((((...))))))... ( -38.00) >consensus CCAGUGGCAAGGCCAAGCCGAGCUGUCAUUGCACCAACAUUAGUAUUAUGCACCUGUUUCAUGAGAUGAAGCAGGAGUUCCCCACGAUACCCGAUGCCAUAGUGACGCAAUGCGUCAAUG ...((((((.(((...)))(((((.....((((...((....))....))))((((((((((...)))))))))))))))..............))))))..((((((...))))))... (-34.96 = -35.32 + 0.36)

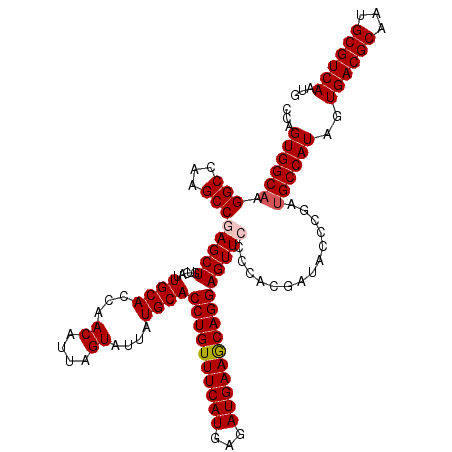
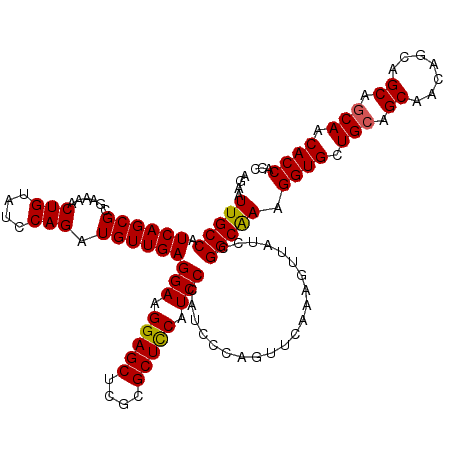
| Location | 14,814,429 – 14,814,549 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -34.80 |
| Energy contribution | -34.60 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14814429 120 + 20766785 AGAAUUGCCAUCAGCGCGAAAACUGUAUCCAGAUGUUGAGGAAGGAGCUCGCGCUUCAUCCUAUCCCAGUGCAGAGCUAUCCGGCAAAGGUGCUGCAGCAACAGCAGCAGCAACACCACC ....(((((.((((((......(((....))).))))))(((...((((((((((............))))).))))).)))))))).((((((((.((....)).)))))...)))... ( -40.40) >DroSec_CAF1 16276 120 + 1 AGAAUUGCCAUCAGCGCGAAAACUGUAUCCAGAUGUUGAGGAAGGAGCUCGCGCUCCAUCCCAUCCCAGUUCAAAGUUAUCCGGCAAAGGUGCUGCAGCAACAGCAGCAACAACACCACC ....(((((.((((((......(((....))).))))))(((.(((((....))))).))).....................))))).(.((((((.......)))))).)......... ( -33.60) >DroSim_CAF1 10549 120 + 1 AGAAUUGCCAUCAGCGCGAAAACUGUAUCCAGAUGUUGAGGAAGGAGCUCGCGCUCCAUCCCAUCCCAGUUCAAAGUUAUCCGGCAAAGGUGCUGCAGCAACAGCAGCAGCAACACCACC ....(((((.((((((......(((....))).))))))(((.(((((....))))).))).....................))))).((((((((.((....)).)))))...)))... ( -37.00) >DroEre_CAF1 9153 120 + 1 AGAAUUGCCAUCAGCGCGAAAACUGUAUCCAGAUGUUGAGGAAGGAGCUUGCGCUCCAUCCCAUCCCAGUUCAAAGUUAUCCGGCGAAGGUGUUGCAGCAACAGCAGCAGCAACACCACC ....(((((.((((((......(((....))).))))))(((.(((((....))))).))).....................))))).((((((((.((.......)).))))))))... ( -39.80) >DroYak_CAF1 10423 120 + 1 AGAAUUGCCAUCAGCGCGAAAACUGUAUCCAGAUGUUGAGGAAGGAGCUCGCGCUCCAUCCCAUCCCAGUUCAAAGUUAUCCGGCGAAGGUGUUGCAGCAACAGCAGCAGCAACACCACC ....(((((.((((((......(((....))).))))))(((.(((((....))))).))).....................))))).((((((((.((.......)).))))))))... ( -39.80) >consensus AGAAUUGCCAUCAGCGCGAAAACUGUAUCCAGAUGUUGAGGAAGGAGCUCGCGCUCCAUCCCAUCCCAGUUCAAAGUUAUCCGGCAAAGGUGCUGCAGCAACAGCAGCAGCAACACCACC ....(((((.((((((......(((....))).))))))(((.(((((....))))).))).....................))))).((((.(((.((.......)).))).))))... (-34.80 = -34.60 + -0.20)

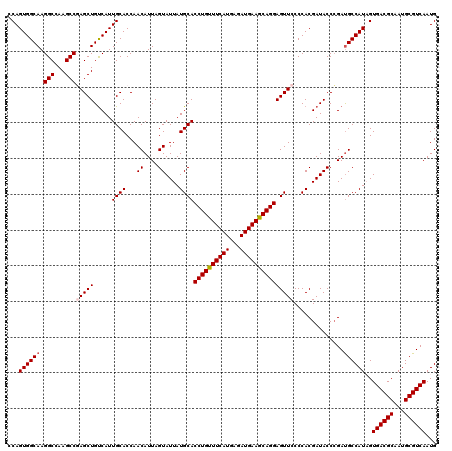
| Location | 14,814,429 – 14,814,549 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -46.74 |
| Consensus MFE | -44.27 |
| Energy contribution | -44.47 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14814429 120 - 20766785 GGUGGUGUUGCUGCUGCUGUUGCUGCAGCACCUUUGCCGGAUAGCUCUGCACUGGGAUAGGAUGAAGCGCGAGCUCCUUCCUCAACAUCUGGAUACAGUUUUCGCGCUGAUGGCAAUUCU ...((((((((.((.......)).)))))))).((((((..((((...((..((((..((((....((....))))))..))))....(((....))).....)))))).)))))).... ( -42.50) >DroSec_CAF1 16276 120 - 1 GGUGGUGUUGUUGCUGCUGUUGCUGCAGCACCUUUGCCGGAUAACUUUGAACUGGGAUGGGAUGGAGCGCGAGCUCCUUCCUCAACAUCUGGAUACAGUUUUCGCGCUGAUGGCAAUUCU ...((((((((.((.......)).)))))))).((((((.........((((((((((((((.(((((....))))).)))....))))).....)))))).........)))))).... ( -46.67) >DroSim_CAF1 10549 120 - 1 GGUGGUGUUGCUGCUGCUGUUGCUGCAGCACCUUUGCCGGAUAACUUUGAACUGGGAUGGGAUGGAGCGCGAGCUCCUUCCUCAACAUCUGGAUACAGUUUUCGCGCUGAUGGCAAUUCU ...((((((((.((.......)).)))))))).((((((.........((((((((((((((.(((((....))))).)))....))))).....)))))).........)))))).... ( -48.27) >DroEre_CAF1 9153 120 - 1 GGUGGUGUUGCUGCUGCUGUUGCUGCAACACCUUCGCCGGAUAACUUUGAACUGGGAUGGGAUGGAGCGCAAGCUCCUUCCUCAACAUCUGGAUACAGUUUUCGCGCUGAUGGCAAUUCU ((.((((((((.((.......)).)))))))).))((((.........((((((((((((((.(((((....))))).)))....))))).....)))))).........))))...... ( -48.37) >DroYak_CAF1 10423 120 - 1 GGUGGUGUUGCUGCUGCUGUUGCUGCAACACCUUCGCCGGAUAACUUUGAACUGGGAUGGGAUGGAGCGCGAGCUCCUUCCUCAACAUCUGGAUACAGUUUUCGCGCUGAUGGCAAUUCU ((.((((((((.((.......)).)))))))).))((((.........((((((((((((((.(((((....))))).)))....))))).....)))))).........))))...... ( -47.87) >consensus GGUGGUGUUGCUGCUGCUGUUGCUGCAGCACCUUUGCCGGAUAACUUUGAACUGGGAUGGGAUGGAGCGCGAGCUCCUUCCUCAACAUCUGGAUACAGUUUUCGCGCUGAUGGCAAUUCU ...((((((((.((.......)).))))))))...((((.........((((((((((((((.(((((....))))).)))....))))).....)))))).........))))...... (-44.27 = -44.47 + 0.20)

| Location | 14,814,469 – 14,814,589 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -38.66 |
| Consensus MFE | -33.58 |
| Energy contribution | -33.34 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14814469 120 + 20766785 GAAGGAGCUCGCGCUUCAUCCUAUCCCAGUGCAGAGCUAUCCGGCAAAGGUGCUGCAGCAACAGCAGCAGCAACACCACCAGAAUAGGCAGGCCAAGCCUCCCACGCCGCUCAAGCCGUC ...((((((((((((............))))).)))))...((((...((((((((.((....)).)))))...))).........((.((((...)))).))..))))......))... ( -41.20) >DroSec_CAF1 16316 120 + 1 GAAGGAGCUCGCGCUCCAUCCCAUCCCAGUUCAAAGUUAUCCGGCAAAGGUGCUGCAGCAACAGCAGCAACAACACCACCAGAAUAGGCAGGCCAAGCCUCCCACGCCGCUCAAGCCGUC ((.(((((....))))).)).....................((((...(.((((((.......)))))).)..........((...(((((((...)))).....)))..))..)))).. ( -33.60) >DroSim_CAF1 10589 120 + 1 GAAGGAGCUCGCGCUCCAUCCCAUCCCAGUUCAAAGUUAUCCGGCAAAGGUGCUGCAGCAACAGCAGCAGCAACACCACCAGAAUAGGCAGGCCAAGCCUCCCACGCCGCUCAAGCCGUC ((.(((((....))))).)).....................((((...((((((((.((....)).)))))...)))....((...(((((((...)))).....)))..))..)))).. ( -37.00) >DroEre_CAF1 9193 120 + 1 GAAGGAGCUUGCGCUCCAUCCCAUCCCAGUUCAAAGUUAUCCGGCGAAGGUGUUGCAGCAACAGCAGCAGCAACACCACCAGAACAGGCAGGCCAAGCCUCCUACGCCCCUUAAGCCGUC ((.(((((....))))).)).....................((((...((((((((.((.......)).))))))))........(((.((((...)))))))...........)))).. ( -40.00) >DroYak_CAF1 10463 120 + 1 GAAGGAGCUCGCGCUCCAUCCCAUCCCAGUUCAAAGUUAUCCGGCGAAGGUGUUGCAGCAACAGCAGCAGCAACACCACCAGAACAGGCAGGCCAAGCCUCCCACGCCGCUCAAACCGUC ((.(((((....))))).)).....................(((((..((((((((.((.......)).)))))))).........((.((((...)))).)).)))))........... ( -41.50) >consensus GAAGGAGCUCGCGCUCCAUCCCAUCCCAGUUCAAAGUUAUCCGGCAAAGGUGCUGCAGCAACAGCAGCAGCAACACCACCAGAAUAGGCAGGCCAAGCCUCCCACGCCGCUCAAGCCGUC ((.(((((....))))).))...............(((...((((...((((.(((.((.......)).))).)))).........((.((((...)))).))..))))....))).... (-33.58 = -33.34 + -0.24)

| Location | 14,814,469 – 14,814,589 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -50.44 |
| Consensus MFE | -48.26 |
| Energy contribution | -47.38 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14814469 120 - 20766785 GACGGCUUGAGCGGCGUGGGAGGCUUGGCCUGCCUAUUCUGGUGGUGUUGCUGCUGCUGUUGCUGCAGCACCUUUGCCGGAUAGCUCUGCACUGGGAUAGGAUGAAGCGCGAGCUCCUUC .(((.(......).)))(((((.((((((...(((((((..((((((((((.((.......)).))))))))...((.(((....)))))))..))))))).....)).))))))))).. ( -49.40) >DroSec_CAF1 16316 120 - 1 GACGGCUUGAGCGGCGUGGGAGGCUUGGCCUGCCUAUUCUGGUGGUGUUGUUGCUGCUGUUGCUGCAGCACCUUUGCCGGAUAACUUUGAACUGGGAUGGGAUGGAGCGCGAGCUCCUUC ...((((.((((..(....)..))))))))..(((((((..((((((((((.((.......)).)))))))).....((((....)))).))..)))))))..(((((....)))))... ( -48.80) >DroSim_CAF1 10589 120 - 1 GACGGCUUGAGCGGCGUGGGAGGCUUGGCCUGCCUAUUCUGGUGGUGUUGCUGCUGCUGUUGCUGCAGCACCUUUGCCGGAUAACUUUGAACUGGGAUGGGAUGGAGCGCGAGCUCCUUC ...((((.((((..(....)..))))))))..(((((((..((((((((((.((.......)).)))))))).....((((....)))).))..)))))))..(((((....)))))... ( -50.40) >DroEre_CAF1 9193 120 - 1 GACGGCUUAAGGGGCGUAGGAGGCUUGGCCUGCCUGUUCUGGUGGUGUUGCUGCUGCUGUUGCUGCAACACCUUCGCCGGAUAACUUUGAACUGGGAUGGGAUGGAGCGCAAGCUCCUUC ..(((.((((((....((((((((...)))).)))).((((((((((((((.((.......)).))))))))...))))))...)))))).)))......((.(((((....))))).)) ( -51.40) >DroYak_CAF1 10463 120 - 1 GACGGUUUGAGCGGCGUGGGAGGCUUGGCCUGCCUGUUCUGGUGGUGUUGCUGCUGCUGUUGCUGCAACACCUUCGCCGGAUAACUUUGAACUGGGAUGGGAUGGAGCGCGAGCUCCUUC ..(((((..((.((((.((.((((...)))).)))))((((((((((((((.((.......)).))))))))...))))))...)))..)))))......((.(((((....))))).)) ( -52.20) >consensus GACGGCUUGAGCGGCGUGGGAGGCUUGGCCUGCCUAUUCUGGUGGUGUUGCUGCUGCUGUUGCUGCAGCACCUUUGCCGGAUAACUUUGAACUGGGAUGGGAUGGAGCGCGAGCUCCUUC ...((((.((((..(....)..))))))))..(((((((..((((((((((.((.......)).)))))))).....((((....)))).))..)))))))..(((((....)))))... (-48.26 = -47.38 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:08 2006