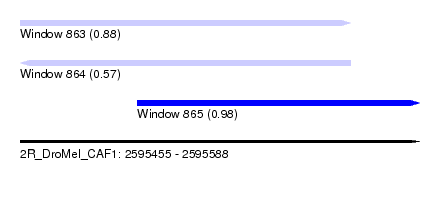
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,595,455 – 2,595,588 |
| Length | 133 |
| Max. P | 0.977412 |

| Location | 2,595,455 – 2,595,565 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 87.18 |
| Mean single sequence MFE | -20.90 |
| Consensus MFE | -16.94 |
| Energy contribution | -17.54 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2595455 110 + 20766785 UAAUUGCCAGAGAAUUUACGAUAAUGUUUUAAAUUAAAAAGGAUGUUGGACCGGGCAUCCCAAGCCCAUCCACAUCCACAACUGUAUCCCUGUCCCCAUGCCCAUUCCCA .......(((.((...((((.((((.......))))....((((((.(((..((((.......)))).))))))))).....)))))).))).................. ( -23.20) >DroSec_CAF1 29628 110 + 1 UAAUUGCCAGAGAAUUUACGAUAAUGUUUUAAAUUAAAAAGGAUGUUGGACCGGGCAUCCCAAGCCCAUCCACAUCCACAACUGUAUCCCUGUCCCCAUGCCCAUCCCCA .......(((.((...((((.((((.......))))....((((((.(((..((((.......)))).))))))))).....)))))).))).................. ( -23.20) >DroSim_CAF1 29154 110 + 1 UAAUUGCCAGAGAAUUUACGAUAAUGUUUUAAAUUAAAAAGGAUGUUGGACCGGGCAUCCCAAGCCCAUCCACAUCCACAACUGUAUCCCUGUCCCCAUGCCCAUCCCCA .......(((.((...((((.((((.......))))....((((((.(((..((((.......)))).))))))))).....)))))).))).................. ( -23.20) >DroEre_CAF1 29469 109 + 1 UAAUUGCCAGAGAAUUUACGAUAAUGUUUUAAAUUAAAAAGGAUGUUGGACCA-GCAACCCAAGCCCAUCCACAUCCACAUCCACAUCCACAUCAACAUCCACAUCCGUA ................((((.((((.......))))....((((((.(((...-((.......))...))))))))).............................)))) ( -15.10) >DroYak_CAF1 28377 94 + 1 UAAUUGCCAGAGAAUUUACGAUAAUGUUUUAAAUUAAAAAGGAUGUUGGACCGGGCAUCCCAAGCCCAUCCACAUCAACAACCGUAACCAUGCC---------------- ...............(((((.((((.......)))).....(((((.(((..((((.......)))).))))))))......))))).......---------------- ( -19.80) >consensus UAAUUGCCAGAGAAUUUACGAUAAUGUUUUAAAUUAAAAAGGAUGUUGGACCGGGCAUCCCAAGCCCAUCCACAUCCACAACUGUAUCCCUGUCCCCAUGCCCAUCCCCA ............((((((.(((...))).)))))).....((((((.(((..((((.......)))).)))))))))................................. (-16.94 = -17.54 + 0.60)

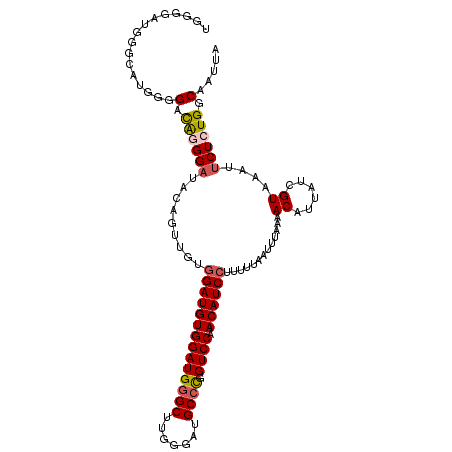
| Location | 2,595,455 – 2,595,565 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 87.18 |
| Mean single sequence MFE | -31.29 |
| Consensus MFE | -22.02 |
| Energy contribution | -22.54 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2595455 110 - 20766785 UGGGAAUGGGCAUGGGGACAGGGAUACAGUUGUGGAUGUGGAUGGGCUUGGGAUGCCCGGUCCAACAUCCUUUUUAAUUUAAAACAUUAUCGUAAAUUCUCUGGCAAUUA .((((((..((.((....))..((((..(((..((((((((((((((.......)))).)))).))))))............)))..))))))..))))))......... ( -33.64) >DroSec_CAF1 29628 110 - 1 UGGGGAUGGGCAUGGGGACAGGGAUACAGUUGUGGAUGUGGAUGGGCUUGGGAUGCCCGGUCCAACAUCCUUUUUAAUUUAAAACAUUAUCGUAAAUUCUCUGGCAAUUA .((((((..((.((....))..((((..(((..((((((((((((((.......)))).)))).))))))............)))..))))))..))))))......... ( -33.14) >DroSim_CAF1 29154 110 - 1 UGGGGAUGGGCAUGGGGACAGGGAUACAGUUGUGGAUGUGGAUGGGCUUGGGAUGCCCGGUCCAACAUCCUUUUUAAUUUAAAACAUUAUCGUAAAUUCUCUGGCAAUUA .((((((..((.((....))..((((..(((..((((((((((((((.......)))).)))).))))))............)))..))))))..))))))......... ( -33.14) >DroEre_CAF1 29469 109 - 1 UACGGAUGUGGAUGUUGAUGUGGAUGUGGAUGUGGAUGUGGAUGGGCUUGGGUUGC-UGGUCCAACAUCCUUUUUAAUUUAAAACAUUAUCGUAAAUUCUCUGGCAAUUA ..((((.((..(((.((((((.((..((((...((((((((((.(((.......))-).)))).))))))..))))..))...)))))).)))..))..))))....... ( -31.60) >DroYak_CAF1 28377 94 - 1 ----------------GGCAUGGUUACGGUUGUUGAUGUGGAUGGGCUUGGGAUGCCCGGUCCAACAUCCUUUUUAAUUUAAAACAUUAUCGUAAAUUCUCUGGCAAUUA ----------------(.((.((((((((((((((((((((((((((.......)))).)))).))))).............))))..)))))))...)).)).)..... ( -24.91) >consensus UGGGGAUGGGCAUGGGGACAGGGAUACAGUUGUGGAUGUGGAUGGGCUUGGGAUGCCCGGUCCAACAUCCUUUUUAAUUUAAAACAUUAUCGUAAAUUCUCUGGCAAUUA ................(.((((((.........((((((((((((((.......)))).)))).)))))).............((......))....)))))).)..... (-22.02 = -22.54 + 0.52)

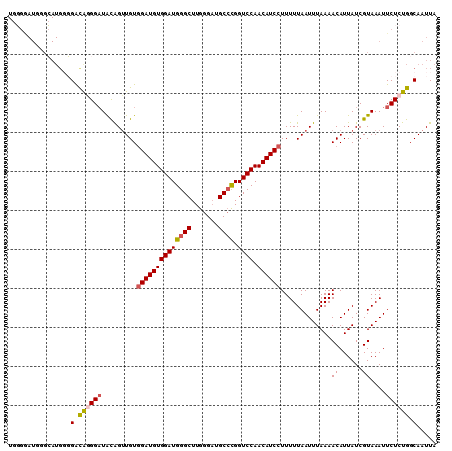
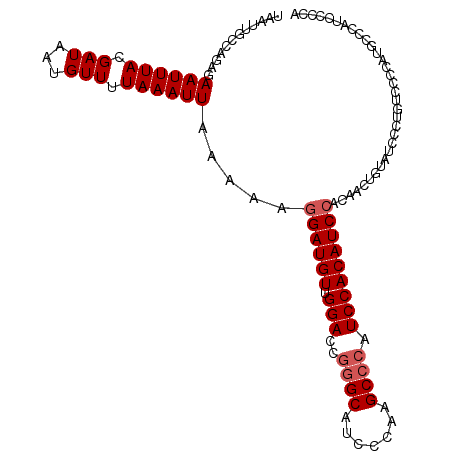
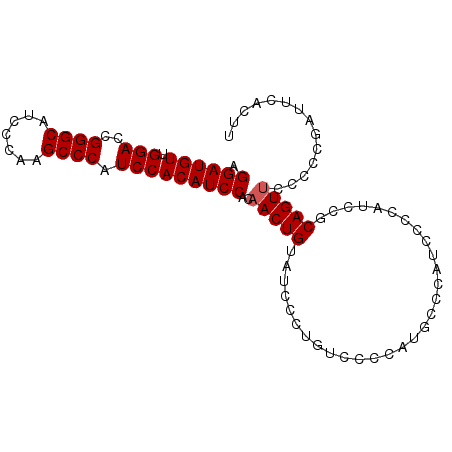
| Location | 2,595,494 – 2,595,588 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 97.87 |
| Mean single sequence MFE | -21.73 |
| Consensus MFE | -20.81 |
| Energy contribution | -21.14 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2595494 94 + 20766785 AGGAUGUUGGACCGGGCAUCCCAAGCCCAUCCACAUCCACAACUGUAUCCCUGUCCCCAUGCCCAUUCCCAUCCUCAGUUCCCCCGAUUCACUU .((((((.(((..((((.......)))).)))))))))..(((((..............................))))).............. ( -21.11) >DroSec_CAF1 29667 94 + 1 AGGAUGUUGGACCGGGCAUCCCAAGCCCAUCCACAUCCACAACUGUAUCCCUGUCCCCAUGCCCAUCCCCAUCCGCAGUUCCCCCGAUUCACUU .((((((.(((..((((.......)))).)))))))))..((((((............................)))))).............. ( -22.49) >DroSim_CAF1 29193 93 + 1 AGGAUGUUGGACCGGGCAUCCCAAGCCCAUCCACAUCCACAACUGUAUCCCUGUCCCCAUGCCCAUCCCCAUCCGCAGU-CCCCCGAUUCACUU .((((((.(((..((((.......)))).)))))))))............((((....(((........)))..)))).-.............. ( -21.60) >consensus AGGAUGUUGGACCGGGCAUCCCAAGCCCAUCCACAUCCACAACUGUAUCCCUGUCCCCAUGCCCAUCCCCAUCCGCAGUUCCCCCGAUUCACUU .((((((.(((..((((.......)))).)))))))))..(((((..............................))))).............. (-20.81 = -21.14 + 0.33)

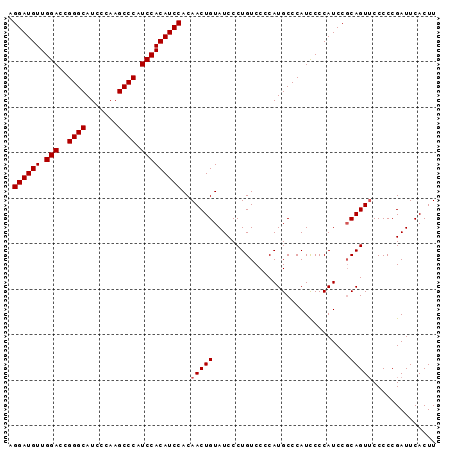
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:56 2006