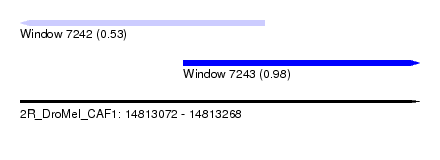
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,813,072 – 14,813,268 |
| Length | 196 |
| Max. P | 0.978021 |

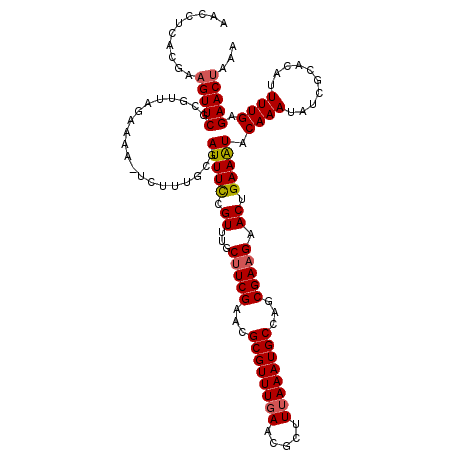
| Location | 14,813,072 – 14,813,192 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.40 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -19.98 |
| Energy contribution | -20.18 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14813072 120 - 20766785 AACCUCACGAAGUUCGCGUUAGAAAAGUCUUUGCAAUUUCCGUUUGCUUCGAACGCGUUUGAACGCUUUAAAUGCCAGCGAAGAACUGAAGUACAAAUAUCGCACAUUUUGAGAACUAAA ..........((((((((..(((....))).))).(((((.((...(((((...((((((((.....))))))))...))))).)).)))))....................)))))... ( -25.90) >DroSec_CAF1 14866 118 - 1 AACCUCACGAAGUUCGCGUUAGAAAA--CUUUGCGAUUUUCGUUUGCUUCGAACGCGUUUGAACGCUUUAAAUGCCAGCGAAGAACUGAAAUACAAAUAUCGCACAUUUUGAGAACUAAA ...((((.((((((..(....)..))--))))((((((((((.((.(((((...((((((((.....))))))))...))))))).))))).......)))))......))))....... ( -28.51) >DroSim_CAF1 9128 118 - 1 AACCUCACGAAGUUCGCGUUAGAAAA--CUUUGCGAUUUUCGUGUGCUUCGAACGCGUUUGAACGCUUUAAAUGCCAGCGAAGAACUGAAAUACAAAUAUCGCACAUUUUGAGAACUAAA ...((((.((((((..(....)..))--))))((((((((((.((.(((((...((((((((.....))))))))...))))).))))))).......)))))......))))....... ( -30.11) >DroEre_CAF1 7748 115 - 1 -----CCCGAAGUUCUCGGUACAAAAAUCAUUUGGAUUUCAGUUUACUUCGAACGCGUUUGAACGCUUUAAAUGCCAGCGAAGAACUGAAAUACAAAUAUCGCACAUUUUGAGAACUAAA -----.....(((((((((..........(((((.(((((((((..(((((...((((((((.....))))))))...)))))))))))))).)))))..........)))))))))... ( -35.65) >DroYak_CAF1 9012 120 - 1 AACCUCUCGAAGUUCUCGUUACAAAAAUCAUUUGGAUUUCAGUUUGCCUCGAACGCGUUUUAACGCUUUAAAUGCCAGCGAAGAACUGAAAUACAAAUAUCGCACAUUUUGAGAACCAAA ...........(((((((...........(((((.((((((((((...(((...((((((.........))))))...))).)))))))))).)))))...........))))))).... ( -29.45) >consensus AACCUCACGAAGUUCGCGUUAGAAAA_UCUUUGCGAUUUCCGUUUGCUUCGAACGCGUUUGAACGCUUUAAAUGCCAGCGAAGAACUGAAAUACAAAUAUCGCACAUUUUGAGAACUAAA ..........(((((....................(((((.((...(((((...((((((((.....))))))))...))))).)).))))).((((..........)))).)))))... (-19.98 = -20.18 + 0.20)

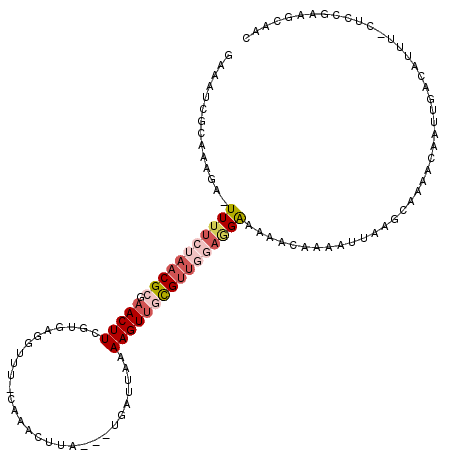
| Location | 14,813,152 – 14,813,268 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.01 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -6.39 |
| Energy contribution | -8.07 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.27 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.978021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14813152 116 + 20766785 GAAAUUGCAAAGACUUUUCUAACGCGAACUUCGUGAGGUUUUCGCACUUA---UGAUUAAAAGUUGCGUUGGAGGAAAAACAAUAUUAAACAAAACAAUUGUCAUUU-CUCCGAAACAAC .......................(((((..((....))..))))).....---.........((((..(((((((((..(((((.............)))))..)))-))))))..)))) ( -27.52) >DroSec_CAF1 14946 115 + 1 AAAAUCGCAAAG--UUUUCUAACGCGAACUUCGUGAGGUUU-CAAGUUUAAAGUAA-AAAAAGUUGCGUUGGAGGAAAAACAAAAUUAAACAAAACAAUUGACAUUU-CUCCGAAGCAAC ....((((.(((--(((.(....).)))))).)))).....-..............-.....(((((.(((((((((...(((...............)))...)))-)))))).))))) ( -25.46) >DroSim_CAF1 9208 116 + 1 AAAAUCGCAAAG--UUUUCUAACGCGAACUUCGUGAGGUUU-CAAGUUUAAAGUGAUUAAAAGUUGCGUUGGAGGAAAAACAAAAUCAAGCAAAACAAUUGUCAUUU-CUCCGAAGCAAC ..((((((.(((--(((.(....).))))))(....)....-..........))))))....(((((.(((((((((..((((...............))))..)))-)))))).))))) ( -29.26) >DroEre_CAF1 7828 97 + 1 GAAAUCCAAAUGAUUUUUGUACCGAGAACUUCGGG--------------------AUUAAAAGUUGUGUUUAAAGGAAAAC--AAUUACGCAAAACAAUUGAAAUUU-GUGCGACGGAAC ....(((..(..(((((((..(((((...))))).--------------------..)))))))..)((((......))))--.....(((...((((.......))-)))))..))).. ( -18.20) >DroYak_CAF1 9092 115 + 1 GAAAUCCAAAUGAUUUUUGUAACGAGAACUUCGAGAGGUUUUCGAACUUA---UGAUUAAAAGUGGUGUAUACCGAACAAA--AAUUACGCAAAACAAUUUAAAUUAAAUACGACCAAAC ..........((((((((((..(((((((((....))))))))).((((.---.......))))(((....)))..)))))--)))))................................ ( -19.20) >consensus GAAAUCGCAAAGA_UUUUCUAACGCGAACUUCGUGAGGUUU_CAAACUUA___UGAUUAAAAGUUGCGUUGGAGGAAAAACAAAAUUAAGCAAAACAAUUGACAUUU_CUCCGAAGCAAC ..............(((((((((((.(((((.............................))))))))))))))))............................................ ( -6.39 = -8.07 + 1.68)

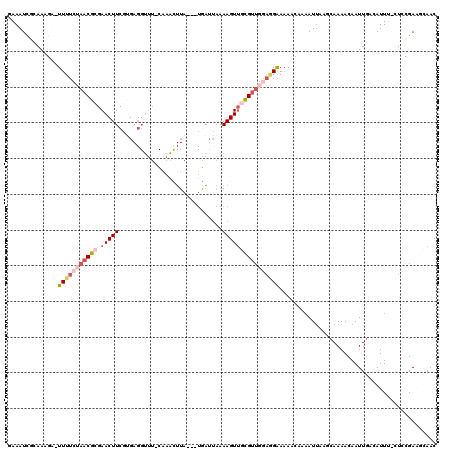
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:01 2006