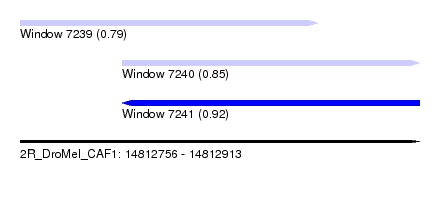
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,812,756 – 14,812,913 |
| Length | 157 |
| Max. P | 0.918946 |

| Location | 14,812,756 – 14,812,873 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.70 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -11.80 |
| Energy contribution | -11.64 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14812756 117 + 20766785 CUAUUAUUUUUCUCACAUUUAUUGCGAAUUUCCCAGCGUAACAACCUACUUGUUCCACAUUGCGUAUGCUUGAUAUGCGCCAUU--UUAUUCAAGUAUUCCCAUUGUGAACUUCACAAA- ............(((((....(((((..........))))).....((((((.........(((((((.....)))))))....--.....)))))).......)))))..........- ( -19.07) >DroSec_CAF1 14584 113 + 1 CUAUUAUUUUUCUCACAUUUAUUUUGAAUUUCCCAGGAUG-UUACCUACUUGUUCCACUAUGCGUAUGCUUAAUAUGCACAAUU--U---UCAAGUGUUCACAUUGUGAACUUCACAAA- ............(((((..(((((((.......)))))))-.....((((((........(((((((.....))))))).....--.---.)))))).......)))))..........- ( -18.96) >DroSim_CAF1 8844 113 + 1 CUAUUAUUUUUCUCACAUUUAUUUUGAAUUUCCCAGGAUG-UUACCUACUUGUUCCACUAUGCGUAUGCUUAAUAUGCACAAUU--U---UCAAGUGUUCACAUUGUGAAGUUCACAAA- ........................((((((((..(.((((-(....((((((........(((((((.....))))))).....--.---.))))))...))))).)))))))))....- ( -21.96) >DroEre_CAF1 7450 115 + 1 CUAUUAUAUUUCUCACAUUUAUUUUGAACUACCCAGCGUAAUAUCCUACUUGUUCCACUAUGCGUAUGCUUAAUAUGCACCACA--U---UCAAGUGCUCACAUUGUGAACUUUAGGAAA ..................((((.(((.......))).))))..(((((...((((((...(((((((.....))))))).(((.--.---....))).......)).))))..))))).. ( -18.50) >DroYak_CAF1 8710 117 + 1 CUAUUAUUUUUCUCACAUUUAUUUUAAACUACCAAGCGUAAUAUCCUACUUGUUCCACGUUGCGUAUGCUUGAUAUCCACUUUGAUA---UCAAGUAUUCACAUUGUGUACUUUAGAAAA .....................(((((((.(((((((((((((................)))))))((((((((((((......))))---)))))))).....))).))).))))))).. ( -24.49) >consensus CUAUUAUUUUUCUCACAUUUAUUUUGAAUUUCCCAGCGUAAUAACCUACUUGUUCCACUAUGCGUAUGCUUAAUAUGCACAAUU__U___UCAAGUGUUCACAUUGUGAACUUCACAAA_ .....................(((((((.(((.(((..........((((((........(((((((.....)))))))............))))))......))).))).))))))).. (-11.80 = -11.64 + -0.16)


| Location | 14,812,796 – 14,812,913 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.44 |
| Mean single sequence MFE | -20.98 |
| Consensus MFE | -10.43 |
| Energy contribution | -10.23 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14812796 117 + 20766785 ACAACCUACUUGUUCCACAUUGCGUAUGCUUGAUAUGCGCCAUU--UUAUUCAAGUAUUCCCAUUGUGAACUUCACAAA-CUUCGCAACGGAAUGUUUUGUUUUUUAUUGUGUGUUUAUU ...........((((.(((.((.(.((((((((...........--....)))))))).).)).))))))).....(((-(..(((((..(((.........)))..))))).))))... ( -22.26) >DroSec_CAF1 14624 99 + 1 -UUACCUACUUGUUCCACUAUGCGUAUGCUUAAUAUGCACAAUU--U---UCAAGUGUUCACAUUGUGAACUUCACAAA-CUUCGCAACGCAAUGUUUUGUUUUUC-------------- -.....((((((........(((((((.....))))))).....--.---.))))))...((((((((...........-........))))))))..........-------------- ( -17.67) >DroSim_CAF1 8884 99 + 1 -UUACCUACUUGUUCCACUAUGCGUAUGCUUAAUAUGCACAAUU--U---UCAAGUGUUCACAUUGUGAAGUUCACAAA-CUUCGCAACGCAAUGUUUUGUUUUUC-------------- -.....((((((........(((((((.....))))))).....--.---.))))))......((((((((((....))-)))))))).((((....)))).....-------------- ( -22.76) >DroEre_CAF1 7490 111 + 1 AUAUCCUACUUGUUCCACUAUGCGUAUGCUUAAUAUGCACCACA--U---UCAAGUGCUCACAUUGUGAACUUUAGGAAACAUCG--ACGAAAUGUUUUGCUUUUCA--GUAUCUUUAUU ...(((((...((((((...(((((((.....))))))).(((.--.---....))).......)).))))..))))).......--..((((.(.....).)))).--........... ( -19.70) >DroYak_CAF1 8750 113 + 1 AUAUCCUACUUGUUCCACGUUGCGUAUGCUUGAUAUCCACUUUGAUA---UCAAGUAUUCACAUUGUGUACUUUAGAAAAAAUGU--ACGAAUUGUUUUGCUUCUCA--GUAUCUUUAUU ...............((((.((.(.((((((((((((......))))---)))))))).).)).))))((((..((((((((((.--......))))))..)))).)--)))........ ( -22.50) >consensus AUAACCUACUUGUUCCACUAUGCGUAUGCUUAAUAUGCACAAUU__U___UCAAGUGUUCACAUUGUGAACUUCACAAA_CUUCGCAACGAAAUGUUUUGUUUUUCA__GU_U_UUUAUU ......((((((........(((((((.....)))))))............))))))......(((((.....))))).......................................... (-10.43 = -10.23 + -0.20)



| Location | 14,812,796 – 14,812,913 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.44 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -8.11 |
| Energy contribution | -8.67 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.35 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14812796 117 - 20766785 AAUAAACACACAAUAAAAAACAAAACAUUCCGUUGCGAAG-UUUGUGAAGUUCACAAUGGGAAUACUUGAAUAA--AAUGGCGCAUAUCAAGCAUACGCAAUGUGGAACAAGUAGGUUGU .........(((((.............((((((((.(((.-((....)).))).)))))))).((((((.....--....(((.((.......)).))).........)))))).))))) ( -22.17) >DroSec_CAF1 14624 99 - 1 --------------GAAAAACAAAACAUUGCGUUGCGAAG-UUUGUGAAGUUCACAAUGUGAACACUUGA---A--AAUUGUGCAUAUUAAGCAUACGCAUAGUGGAACAAGUAGGUAA- --------------.....(((((...((((...))))..-)))))...((((((...))))))(((((.---.--.(((((((.(((.....))).)))))))....)))))......- ( -21.60) >DroSim_CAF1 8884 99 - 1 --------------GAAAAACAAAACAUUGCGUUGCGAAG-UUUGUGAACUUCACAAUGUGAACACUUGA---A--AAUUGUGCAUAUUAAGCAUACGCAUAGUGGAACAAGUAGGUAA- --------------.............(..(((((.((((-((....)))))).)))))..)..(((((.---.--.(((((((.(((.....))).)))))))....)))))......- ( -25.30) >DroEre_CAF1 7490 111 - 1 AAUAAAGAUAC--UGAAAAGCAAAACAUUUCGU--CGAUGUUUCCUAAAGUUCACAAUGUGAGCACUUGA---A--UGUGGUGCAUAUUAAGCAUACGCAUAGUGGAACAAGUAGGAUAU ..........(--(....))...((((((....--.))))))(((((...(((((.(((((..(((....---.--.)))((((.......)))).))))).))))).....)))))... ( -24.80) >DroYak_CAF1 8750 113 - 1 AAUAAAGAUAC--UGAGAAGCAAAACAAUUCGU--ACAUUUUUUCUAAAGUACACAAUGUGAAUACUUGA---UAUCAAAGUGGAUAUCAAGCAUACGCAACGUGGAACAAGUAGGAUAU ........(((--(((((((....((.....))--....))))))....((.(((..((((.((.(((((---((((......))))))))).)).))))..)))..)).))))...... ( -22.00) >consensus AAUAAA_A_AC__UGAAAAACAAAACAUUGCGUUGCGAAG_UUUGUGAAGUUCACAAUGUGAACACUUGA___A__AAUGGUGCAUAUUAAGCAUACGCAUAGUGGAACAAGUAGGUUAU ..................................................(((((.(((((....(((((.................)))))....))))).)))))............. ( -8.11 = -8.67 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:59 2006