
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,812,562 – 14,812,716 |
| Length | 154 |
| Max. P | 0.998697 |

| Location | 14,812,562 – 14,812,679 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.29 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -25.53 |
| Energy contribution | -24.87 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.54 |
| Structure conservation index | 1.04 |
| SVM decision value | 3.19 |
| SVM RNA-class probability | 0.998697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14812562 117 + 20766785 GCUUCUUGUUGUUGUUGC---CUUUUUUCGAAUAUUUCAUUCAAUUUUCUUAGCUUCGUAGUGUUGGUGUGAAAAACACCCACACAAAAAUUUAUUUUUAUAAAAGAGAGCGGCUAAGAG ...(((((.....(((((---(((((((.((((.....))))..................((((.(((((.....))))).))))................))))))).)))))))))). ( -25.60) >DroEre_CAF1 7257 116 + 1 GCU--UUCUUGUUGUUGCUGCGCUUUUUCGAAUAUUUCAUUCAAUUUUCUUAGCUUCGCAGUGUUGGUGUGA--AACACCCACACAAAAAUUUAUUUUUAUAAAAGAGAGCGGCUAAGAG ...--.(((((.....(((((.((((((.((((.....))))..................((((.((((...--..)))).)))).................)))))).)))))))))). ( -26.20) >DroYak_CAF1 8536 97 + 1 ---------------------UAUUUUUCGAAUAUUUCAUUCAAUUUUCUUGGCUUUGUAGUGUUGGUGUGA--AACACCCACACAAAAAUUUAUUUUUAUAAAAGAGAGCGGCUAAGAG ---------------------........((((.....)))).....((((((((.....((((.((((...--..)))).)))).....(((((....))))).......)))))))). ( -22.10) >consensus GCU__UU_UUGUUGUUGC___CAUUUUUCGAAUAUUUCAUUCAAUUUUCUUAGCUUCGUAGUGUUGGUGUGA__AACACCCACACAAAAAUUUAUUUUUAUAAAAGAGAGCGGCUAAGAG .............................((((.....)))).....(((((((..(((.((((.(((((.....))))).)))).....(((((....))))).....)))))))))). (-25.53 = -24.87 + -0.66)

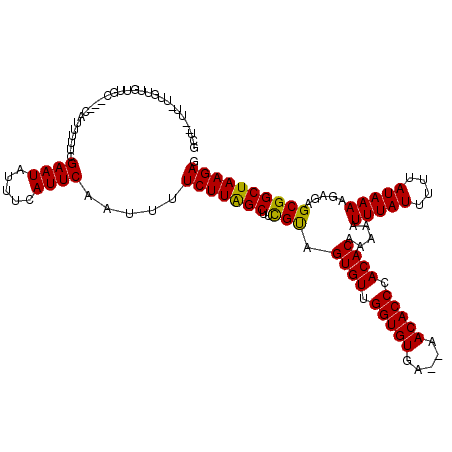
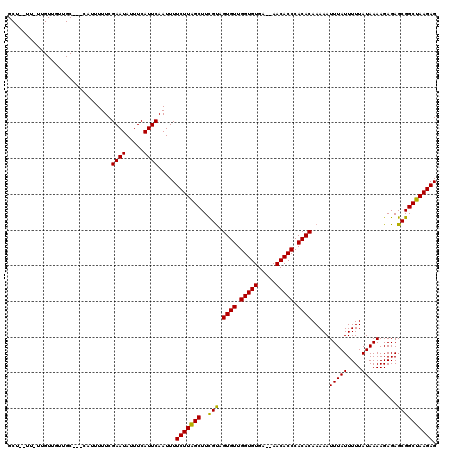
| Location | 14,812,562 – 14,812,679 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.29 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -20.33 |
| Energy contribution | -20.67 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14812562 117 - 20766785 CUCUUAGCCGCUCUCUUUUAUAAAAAUAAAUUUUUGUGUGGGUGUUUUUCACACCAACACUACGAAGCUAAGAAAAUUGAAUGAAAUAUUCGAAAAAAG---GCAACAACAACAAGAAGC .(((((((........(((((....))))).....((((.(((((.....))))).))))......)))))))...(((((((...))))))).....(---....)............. ( -23.00) >DroEre_CAF1 7257 116 - 1 CUCUUAGCCGCUCUCUUUUAUAAAAAUAAAUUUUUGUGUGGGUGUU--UCACACCAACACUGCGAAGCUAAGAAAAUUGAAUGAAAUAUUCGAAAAAGCGCAGCAACAACAAGAA--AGC .((((((((((.....(((((....))))).....((((.((((..--...)))).)))).)))..)))))))...(((((((...)))))))......................--... ( -24.60) >DroYak_CAF1 8536 97 - 1 CUCUUAGCCGCUCUCUUUUAUAAAAAUAAAUUUUUGUGUGGGUGUU--UCACACCAACACUACAAAGCCAAGAAAAUUGAAUGAAAUAUUCGAAAAAUA--------------------- .((((.((........(((((....))))).....((((.((((..--...)))).))))......)).))))...(((((((...)))))))......--------------------- ( -16.90) >consensus CUCUUAGCCGCUCUCUUUUAUAAAAAUAAAUUUUUGUGUGGGUGUU__UCACACCAACACUACGAAGCUAAGAAAAUUGAAUGAAAUAUUCGAAAAAAA___GCAACAACAA_AA__AGC .(((((((........(((((....))))).....((((.(((((.....))))).))))......)))))))...(((((((...)))))))........................... (-20.33 = -20.67 + 0.33)

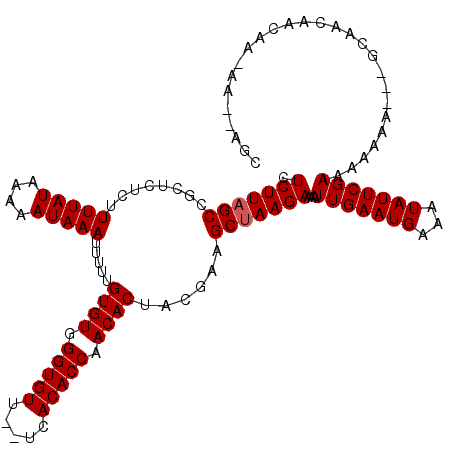
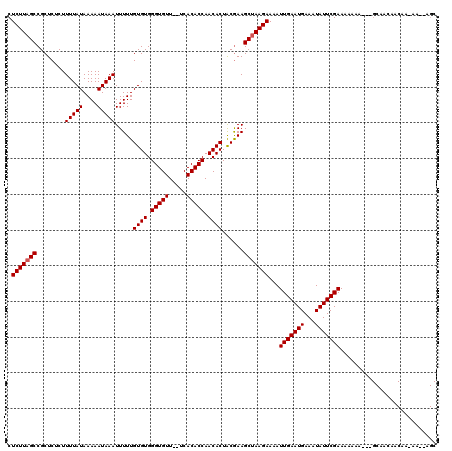
| Location | 14,812,599 – 14,812,716 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -25.22 |
| Energy contribution | -25.14 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14812599 117 + 20766785 UCAAUUUUCUUAGCUUCGUAGUGUUGGUGUGAAAAACACCCACACAAAAAUUUAUUUUUAUAAAAGAGAGCGGCUAAGAGAAAGAUCCAAAUUUCUG---UUACAAAGUGGUAUUGGUGU ....(((((((((((.(...((((.(((((.....))))).)))).....(((((....))))).....).)))))))))))....((((((..((.---......))..)).))))... ( -29.80) >DroSec_CAF1 14424 120 + 1 UCAAUUUUCUUAGCUUCGUAGUAUUGGUUUGAAAAACACCCACACAAAAAUUUAUUUUUAUAAAAGAGAGCGGCUAAGAGAAAGAUCCAAAUUUCUGUUGUUACAAAGUGGUAUUGGUGU ....(((((((((((.(...((...(((((....)).)))...)).....(((((....))))).....).)))))))))))....((((....(..((.......))..)..))))... ( -19.30) >DroSim_CAF1 8684 120 + 1 UCAAUUUUCUUAGCUUCGUAGUGUUGGUGUGAAAAACACCCACACAAAAAUUUAUUUUUAUAAAAGAGAGCGGCUAAGAGAAAGAUCCAAAUUUCUGUUGUUACAAAGUGGUAUUGGUGU ....(((((((((((.(...((((.(((((.....))))).)))).....(((((....))))).....).)))))))))))....((((....(..((.......))..)..))))... ( -30.00) >DroEre_CAF1 7295 115 + 1 UCAAUUUUCUUAGCUUCGCAGUGUUGGUGUGA--AACACCCACACAAAAAUUUAUUUUUAUAAAAGAGAGCGGCUAAGAGAAAGAUACAAAUUUCUU---UUACAAAGUGGUAUUAGUGU ....((((((((((..(((.((((.((((...--..)))).)))).....(((((....))))).....))))))))))))).(((((...(((...---....)))...)))))..... ( -29.00) >DroYak_CAF1 8555 115 + 1 UCAAUUUUCUUGGCUUUGUAGUGUUGGUGUGA--AACACCCACACAAAAAUUUAUUUUUAUAAAAGAGAGCGGCUAAGAGAAAGAUCGAAAUUUCUU---UUACAAAGUGGUAUUGGUGU (((((.....(.((((((((((((.((((...--..)))).)))).................(((((((.(((((.......)).)))...))))))---))))))))).).)))))... ( -27.40) >consensus UCAAUUUUCUUAGCUUCGUAGUGUUGGUGUGAAAAACACCCACACAAAAAUUUAUUUUUAUAAAAGAGAGCGGCUAAGAGAAAGAUCCAAAUUUCUG___UUACAAAGUGGUAUUGGUGU ....((((((((((..(((.((((.(((((.....))))).)))).....(((((....))))).....)))))))))))))(((........)))........................ (-25.22 = -25.14 + -0.08)

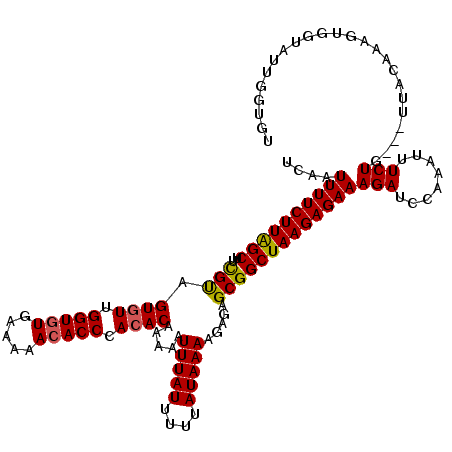
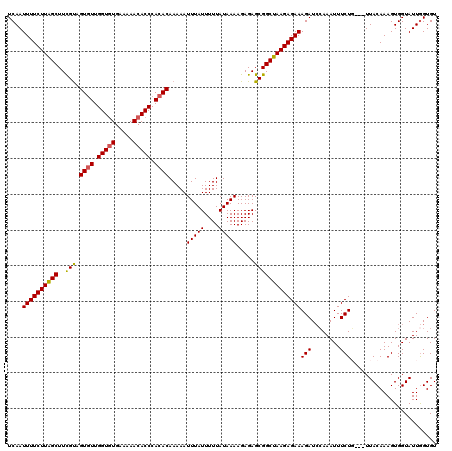
| Location | 14,812,599 – 14,812,716 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -18.58 |
| Energy contribution | -18.66 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14812599 117 - 20766785 ACACCAAUACCACUUUGUAA---CAGAAAUUUGGAUCUUUCUCUUAGCCGCUCUCUUUUAUAAAAAUAAAUUUUUGUGUGGGUGUUUUUCACACCAACACUACGAAGCUAAGAAAAUUGA ....((((.(((..((....---....))..))).......(((((((........(((((....))))).....((((.(((((.....))))).))))......)))))))..)))). ( -23.00) >DroSec_CAF1 14424 120 - 1 ACACCAAUACCACUUUGUAACAACAGAAAUUUGGAUCUUUCUCUUAGCCGCUCUCUUUUAUAAAAAUAAAUUUUUGUGUGGGUGUUUUUCAAACCAAUACUACGAAGCUAAGAAAAUUGA ....((((.(((.(((((....)))))....))).......((((((((((((.....((((((((....)))))))).)))))...(((.............))))))))))..)))). ( -19.12) >DroSim_CAF1 8684 120 - 1 ACACCAAUACCACUUUGUAACAACAGAAAUUUGGAUCUUUCUCUUAGCCGCUCUCUUUUAUAAAAAUAAAUUUUUGUGUGGGUGUUUUUCACACCAACACUACGAAGCUAAGAAAAUUGA ....((((.(((.(((((....)))))....))).......(((((((........(((((....))))).....((((.(((((.....))))).))))......)))))))..)))). ( -24.10) >DroEre_CAF1 7295 115 - 1 ACACUAAUACCACUUUGUAA---AAGAAAUUUGUAUCUUUCUCUUAGCCGCUCUCUUUUAUAAAAAUAAAUUUUUGUGUGGGUGUU--UCACACCAACACUGCGAAGCUAAGAAAAUUGA ...................(---((((........))))).((((((((((.....(((((....))))).....((((.((((..--...)))).)))).)))..)))))))....... ( -23.00) >DroYak_CAF1 8555 115 - 1 ACACCAAUACCACUUUGUAA---AAGAAAUUUCGAUCUUUCUCUUAGCCGCUCUCUUUUAUAAAAAUAAAUUUUUGUGUGGGUGUU--UCACACCAACACUACAAAGCCAAGAAAAUUGA ....((((.....(((((((---((((.....((..((.......)).))...))))))))))).......(((((((((((((..--...))))..))).))))))........)))). ( -18.30) >consensus ACACCAAUACCACUUUGUAA___CAGAAAUUUGGAUCUUUCUCUUAGCCGCUCUCUUUUAUAAAAAUAAAUUUUUGUGUGGGUGUUUUUCACACCAACACUACGAAGCUAAGAAAAUUGA ....((((.................((((........))))(((((((........(((((....))))).....((((.(((((.....))))).))))......)))))))..)))). (-18.58 = -18.66 + 0.08)

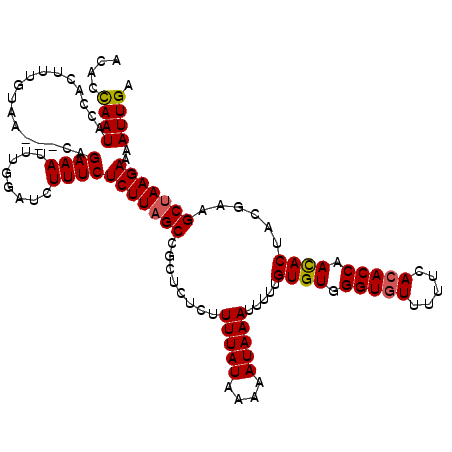
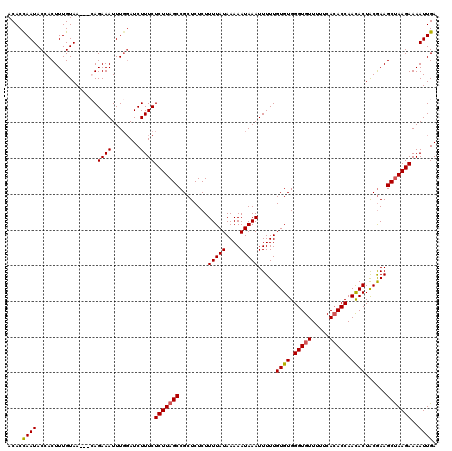
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:56 2006