| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,812,208 – 14,812,328 |
| Length | 120 |
| Max. P | 0.799586 |

| Location | 14,812,208 – 14,812,328 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -17.93 |
| Energy contribution | -17.33 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
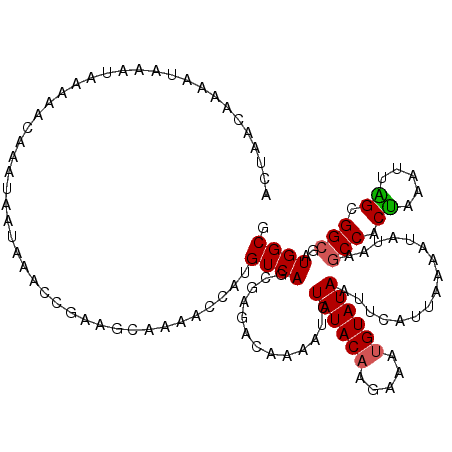
>2R_DroMel_CAF1 14812208 120 + 20766785 CGCCAUCGCCGCUAAUUUAGUGGCUUAUAUUUGAAUGAAUUAUACAUUUCUUGUAUAGAUUUUGUCUCGCUGACAUGGUUUUGCUUCGGUUUAUUAUUUGUUUUUAUUUAUUUUGUUAGU .......(((((((...))))))).......(((((((((((((((.....)))))))....((((.....))))((((...((....))..))))......)))))))).......... ( -23.20) >DroSec_CAF1 14049 114 + 1 CGCCAUCGCCGCUAAUUUAGUGGCUUAUAUUUUAAUGAAUUAUACAUUUCUUGUAUAGAUUUUGUCUCGCUGACAUGGUUUUG------UUUAUUAUUUGUUUUUAUUUAUUUUGUUAGU .......(((((((...)))))))........((((((((((((((.....))))))(((..((((.....))))..)))..)------)))))))........................ ( -21.70) >DroSim_CAF1 8311 120 + 1 CGCCAUCGCCGCUAAUUUAGUGGCUUAUAUUUUAAUGAAUUAUACAUUUCUUGUAUAGAUUUUGUCGCGCUGACAUGGUUUUGCUUCGGUUUAUUAUUUGUUUUUAUUUAUUUUGUUAGU .((....(((((((...)))))))...............(((((((.....)))))))........))(((((((..((...((....))...................))..))))))) ( -22.95) >DroEre_CAF1 6901 119 + 1 CGCCAUCGCCGCCUAUUUAGUGGGUUAUAUUUUAAUAAAUUAUACGUU-UUUGUAUAGAUUUUGUGUCUCUGACAUGGUUUUGCUUCGCUUUAUUAUUUGUUUUUAUUUAUUUUGUCAGA (((.(((...((((((...))))))...............((((((..-..)))))))))...)))..(((((((..((........((..........))........))..))))))) ( -17.09) >DroYak_CAF1 8204 120 + 1 CGCCAUCGCCGCCAAUUGGGUGGCUUAUAUUUUAAUGAAUUAUACAUUUUUUGUAUAGAUUUCGUGUCGCUGACAUGGUUUUGCUUCGCUUUAUUAUUUGUUUUUAUUUAUUUUGUCAGA .......((((((.....))))))..........((((((((((((.....)))))))..)))))....((((((..((........((..........))........))..)))))). ( -25.29) >consensus CGCCAUCGCCGCUAAUUUAGUGGCUUAUAUUUUAAUGAAUUAUACAUUUCUUGUAUAGAUUUUGUCUCGCUGACAUGGUUUUGCUUCGCUUUAUUAUUUGUUUUUAUUUAUUUUGUUAGU .......((((((.....))))))..........((((((((((((.....)))))))..)))))...(((((((..((..............................))..))))))) (-17.93 = -17.33 + -0.60)


| Location | 14,812,208 – 14,812,328 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -14.98 |
| Consensus MFE | -11.56 |
| Energy contribution | -11.56 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14812208 120 - 20766785 ACUAACAAAAUAAAUAAAAACAAAUAAUAAACCGAAGCAAAACCAUGUCAGCGAGACAAAAUCUAUACAAGAAAUGUAUAAUUCAUUCAAAUAUAAGCCACUAAAUUAGCGGCGAUGGCG ....................................((.......((((.....))))..(((((((((.....))))))................(((.(((...))).)))))).)). ( -15.80) >DroSec_CAF1 14049 114 - 1 ACUAACAAAAUAAAUAAAAACAAAUAAUAAA------CAAAACCAUGUCAGCGAGACAAAAUCUAUACAAGAAAUGUAUAAUUCAUUAAAAUAUAAGCCACUAAAUUAGCGGCGAUGGCG ...............................------.....((.((((.....))))..(((((((((.....))))))................(((.(((...))).)))))))).. ( -14.70) >DroSim_CAF1 8311 120 - 1 ACUAACAAAAUAAAUAAAAACAAAUAAUAAACCGAAGCAAAACCAUGUCAGCGCGACAAAAUCUAUACAAGAAAUGUAUAAUUCAUUAAAAUAUAAGCCACUAAAUUAGCGGCGAUGGCG ....................................((.......((((.....))))..(((((((((.....))))))................(((.(((...))).)))))).)). ( -15.80) >DroEre_CAF1 6901 119 - 1 UCUGACAAAAUAAAUAAAAACAAAUAAUAAAGCGAAGCAAAACCAUGUCAGAGACACAAAAUCUAUACAAA-AACGUAUAAUUUAUUAAAAUAUAACCCACUAAAUAGGCGGCGAUGGCG (((((((......................................)))))))........((((((((...-...))))).................((.((.....)).)).))).... ( -10.66) >DroYak_CAF1 8204 120 - 1 UCUGACAAAAUAAAUAAAAACAAAUAAUAAAGCGAAGCAAAACCAUGUCAGCGACACGAAAUCUAUACAAAAAAUGUAUAAUUCAUUAAAAUAUAAGCCACCCAAUUGGCGGCGAUGGCG .((((((......................................)))))).....((..(((((((((.....))))))................(((.((.....)).))))))..)) ( -17.96) >consensus ACUAACAAAAUAAAUAAAAACAAAUAAUAAACCGAAGCAAAACCAUGUCAGCGAGACAAAAUCUAUACAAGAAAUGUAUAAUUCAUUAAAAUAUAAGCCACUAAAUUAGCGGCGAUGGCG ..............................................((((.............((((((.....))))))................(((.((.....)).)))..)))). (-11.56 = -11.56 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:51 2006