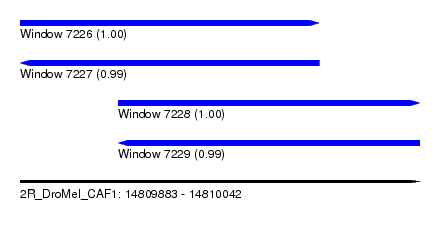
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,809,883 – 14,810,042 |
| Length | 159 |
| Max. P | 0.999547 |

| Location | 14,809,883 – 14,810,002 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.49 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -30.84 |
| Energy contribution | -30.84 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14809883 119 + 20766785 GCCCCUCCGAGUUUGAU-UCCCAUGCCCAUAUUCAAUUCUAGAAUUCGGCAAUGACGUCAAGGCCUGACGUCUGCAGAAAAUUCCGCAAAACGCCUCUUCAAAUUAUUGGCGAGGCAAGA (((.(.((((((((((.-.....((((...((((.......))))..))))..(((((((.....)))))))(((.((....)).)))..........))))))..)))).).))).... ( -32.80) >DroSec_CAF1 11719 119 + 1 GCCCCUCCGAGUUUGAU-UCCCAUGCCCAUAUUCAAUUCUAGAAUUCGGCAAUGACGUCAAGGCCUGACGUCUGCAGAAAAUUCCGCAAAACGCCUCUUCAAAUUAUUGGCGAGGCAAGA (((.(.((((((((((.-.....((((...((((.......))))..))))..(((((((.....)))))))(((.((....)).)))..........))))))..)))).).))).... ( -32.80) >DroSim_CAF1 6004 119 + 1 GCCCCUCCGAGUUUGAU-UCCCAUGCCCAUAUUCAAUUCUAGAAUUCGGCAAUGACGUCAAGGCCUGACGUCUGCAGAAAAUUCCGCAAAACGCCUCUUCAAAUUAUUGGCGAGGCAAGA (((.(.((((((((((.-.....((((...((((.......))))..))))..(((((((.....)))))))(((.((....)).)))..........))))))..)))).).))).... ( -32.80) >DroEre_CAF1 4550 120 + 1 CCCCCACCGAUUUUGAUUUCCCAUGCCCAUAUUCUAUUCUAGAAUUCGGCAAUGACGUCAAGGCCUGACGUCUGCAGAAAAUUCCGCAAAACGCCUCUUCAAAUUAUUGGCGAGGCAAGA .......................((((...((((((...))))))..))))..(((((((.....)))))))(((.((....)).)))....(((((.((((....)))).))))).... ( -32.40) >DroYak_CAF1 5774 120 + 1 UCCCCGCCGAUUUUGAUUUCCCAUGCCCAUAUUCAAUUCUAGAAUUCGGCAAUGACGUCAAGGCCUGACGUCUGCAGAAAAUUCCGCAAAACGCCUCUUCAAAUUAUUGGCGAGGCAAGA ..((((((((((((((.......((((...((((.......))))..))))..(((((((.....)))))))(((.((....)).)))..........)))))..))))))).))..... ( -33.70) >consensus GCCCCUCCGAGUUUGAU_UCCCAUGCCCAUAUUCAAUUCUAGAAUUCGGCAAUGACGUCAAGGCCUGACGUCUGCAGAAAAUUCCGCAAAACGCCUCUUCAAAUUAUUGGCGAGGCAAGA .......................((((...((((.......))))..))))..(((((((.....)))))))(((.((....)).)))....(((((.((((....)))).))))).... (-30.84 = -30.84 + -0.00)


| Location | 14,809,883 – 14,810,002 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.49 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -36.08 |
| Energy contribution | -36.48 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14809883 119 - 20766785 UCUUGCCUCGCCAAUAAUUUGAAGAGGCGUUUUGCGGAAUUUUCUGCAGACGUCAGGCCUUGACGUCAUUGCCGAAUUCUAGAAUUGAAUAUGGGCAUGGGA-AUCAAACUCGGAGGGGC ....(((((.((.............(((((((.(((((....)))))))))))).((..((((..((..((((.(((((.......)))).).))))..)).-.))))..)))).))))) ( -38.10) >DroSec_CAF1 11719 119 - 1 UCUUGCCUCGCCAAUAAUUUGAAGAGGCGUUUUGCGGAAUUUUCUGCAGACGUCAGGCCUUGACGUCAUUGCCGAAUUCUAGAAUUGAAUAUGGGCAUGGGA-AUCAAACUCGGAGGGGC ....(((((.((.............(((((((.(((((....)))))))))))).((..((((..((..((((.(((((.......)))).).))))..)).-.))))..)))).))))) ( -38.10) >DroSim_CAF1 6004 119 - 1 UCUUGCCUCGCCAAUAAUUUGAAGAGGCGUUUUGCGGAAUUUUCUGCAGACGUCAGGCCUUGACGUCAUUGCCGAAUUCUAGAAUUGAAUAUGGGCAUGGGA-AUCAAACUCGGAGGGGC ....(((((.((.............(((((((.(((((....)))))))))))).((..((((..((..((((.(((((.......)))).).))))..)).-.))))..)))).))))) ( -38.10) >DroEre_CAF1 4550 120 - 1 UCUUGCCUCGCCAAUAAUUUGAAGAGGCGUUUUGCGGAAUUUUCUGCAGACGUCAGGCCUUGACGUCAUUGCCGAAUUCUAGAAUAGAAUAUGGGCAUGGGAAAUCAAAAUCGGUGGGGG .....(((((((.............(((((((.(((((....)))))))))))).((..((((..((..((((.(((((((...)))))).).))))...))..))))..))))))))). ( -42.50) >DroYak_CAF1 5774 120 - 1 UCUUGCCUCGCCAAUAAUUUGAAGAGGCGUUUUGCGGAAUUUUCUGCAGACGUCAGGCCUUGACGUCAUUGCCGAAUUCUAGAAUUGAAUAUGGGCAUGGGAAAUCAAAAUCGGCGGGGA .....(((((((.............(((((((.(((((....)))))))))))).((..((((..((..((((.(((((.......)))).).))))...))..))))..))))))))). ( -43.10) >consensus UCUUGCCUCGCCAAUAAUUUGAAGAGGCGUUUUGCGGAAUUUUCUGCAGACGUCAGGCCUUGACGUCAUUGCCGAAUUCUAGAAUUGAAUAUGGGCAUGGGA_AUCAAACUCGGAGGGGC ....(((((.((.............(((((((.(((((....)))))))))))).((..((((..((..((((.(((((.......)))).).))))...))..))))..)))).))))) (-36.08 = -36.48 + 0.40)


| Location | 14,809,922 – 14,810,042 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -31.94 |
| Consensus MFE | -30.78 |
| Energy contribution | -30.78 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.71 |
| SVM RNA-class probability | 0.999547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14809922 120 + 20766785 AGAAUUCGGCAAUGACGUCAAGGCCUGACGUCUGCAGAAAAUUCCGCAAAACGCCUCUUCAAAUUAUUGGCGAGGCAAGAAAAGCAAAAGAAAAUCGGCGAGCGAGCAGCGAAAUACAAA .(((((..(((..(((((((.....))))))))))....)))))(((.....(((((.((((....)))).))))).....................((......)).)))......... ( -32.20) >DroSec_CAF1 11758 120 + 1 AGAAUUCGGCAAUGACGUCAAGGCCUGACGUCUGCAGAAAAUUCCGCAAAACGCCUCUUCAAAUUAUUGGCGAGGCAAGAAAAGCAAAAGAAAAUCGGCGAGCGAGCAGCGAAAUACAAA .(((((..(((..(((((((.....))))))))))....)))))(((.....(((((.((((....)))).))))).....................((......)).)))......... ( -32.20) >DroSim_CAF1 6043 120 + 1 AGAAUUCGGCAAUGACGUCAAGGCCUGACGUCUGCAGAAAAUUCCGCAAAACGCCUCUUCAAAUUAUUGGCGAGGCAAGAAAAGCAAAAGAAAAUCGGCGAGCGAGCAGCGAAAUACAAA .(((((..(((..(((((((.....))))))))))....)))))(((.....(((((.((((....)))).))))).....................((......)).)))......... ( -32.20) >DroEre_CAF1 4590 120 + 1 AGAAUUCGGCAAUGACGUCAAGGCCUGACGUCUGCAGAAAAUUCCGCAAAACGCCUCUUCAAAUUAUUGGCGAGGCAAGAAAAGCUAAAGAAAAUCGGCGAGCGAGCACCAGAAUACAAA .(.((((.(((..(((((((.....)))))))))).........(((.....(((((.((((....)))).))))).......((..(......)..))..))).......)))).)... ( -32.10) >DroYak_CAF1 5814 120 + 1 AGAAUUCGGCAAUGACGUCAAGGCCUGACGUCUGCAGAAAAUUCCGCAAAACGCCUCUUCAAAUUAUUGGCGAGGCAAGAAAAGCAAAAGAAAAUCGGCGAGCGAGCACCAAAAUACAAA .(((((..(((..(((((((.....))))))))))....)))))(((.....(((((.((((....)))).))))).......((....(.....).))..)))................ ( -31.00) >consensus AGAAUUCGGCAAUGACGUCAAGGCCUGACGUCUGCAGAAAAUUCCGCAAAACGCCUCUUCAAAUUAUUGGCGAGGCAAGAAAAGCAAAAGAAAAUCGGCGAGCGAGCAGCGAAAUACAAA .(((((..(((..(((((((.....))))))))))....)))))(((.....(((((.((((....)))).))))).......((....(.....).))..)))................ (-30.78 = -30.78 + -0.00)



| Location | 14,809,922 – 14,810,042 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -30.82 |
| Energy contribution | -30.82 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14809922 120 - 20766785 UUUGUAUUUCGCUGCUCGCUCGCCGAUUUUCUUUUGCUUUUCUUGCCUCGCCAAUAAUUUGAAGAGGCGUUUUGCGGAAUUUUCUGCAGACGUCAGGCCUUGACGUCAUUGCCGAAUUCU .....(((((((.((......((.((......)).)).......(((((..(((....)))..)))))))...))))))).(((.(((((((((((...))))))))..))).))).... ( -32.80) >DroSec_CAF1 11758 120 - 1 UUUGUAUUUCGCUGCUCGCUCGCCGAUUUUCUUUUGCUUUUCUUGCCUCGCCAAUAAUUUGAAGAGGCGUUUUGCGGAAUUUUCUGCAGACGUCAGGCCUUGACGUCAUUGCCGAAUUCU .....(((((((.((......((.((......)).)).......(((((..(((....)))..)))))))...))))))).(((.(((((((((((...))))))))..))).))).... ( -32.80) >DroSim_CAF1 6043 120 - 1 UUUGUAUUUCGCUGCUCGCUCGCCGAUUUUCUUUUGCUUUUCUUGCCUCGCCAAUAAUUUGAAGAGGCGUUUUGCGGAAUUUUCUGCAGACGUCAGGCCUUGACGUCAUUGCCGAAUUCU .....(((((((.((......((.((......)).)).......(((((..(((....)))..)))))))...))))))).(((.(((((((((((...))))))))..))).))).... ( -32.80) >DroEre_CAF1 4590 120 - 1 UUUGUAUUCUGGUGCUCGCUCGCCGAUUUUCUUUAGCUUUUCUUGCCUCGCCAAUAAUUUGAAGAGGCGUUUUGCGGAAUUUUCUGCAGACGUCAGGCCUUGACGUCAUUGCCGAAUUCU ...(.(((((((((......)))))..........((.......(((((..(((....)))..)))))....((((((....))))))((((((((...))))))))...)).)))).). ( -33.40) >DroYak_CAF1 5814 120 - 1 UUUGUAUUUUGGUGCUCGCUCGCCGAUUUUCUUUUGCUUUUCUUGCCUCGCCAAUAAUUUGAAGAGGCGUUUUGCGGAAUUUUCUGCAGACGUCAGGCCUUGACGUCAUUGCCGAAUUCU ........((((((......)))))).....((((((......((((((..(((....)))..))))))....))))))..(((.(((((((((((...))))))))..))).))).... ( -34.10) >consensus UUUGUAUUUCGCUGCUCGCUCGCCGAUUUUCUUUUGCUUUUCUUGCCUCGCCAAUAAUUUGAAGAGGCGUUUUGCGGAAUUUUCUGCAGACGUCAGGCCUUGACGUCAUUGCCGAAUUCU .....(((((((.((......((.((......)).)).......(((((..(((....)))..)))))))...))))))).(((.(((((((((((...))))))))..))).))).... (-30.82 = -30.82 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:47 2006