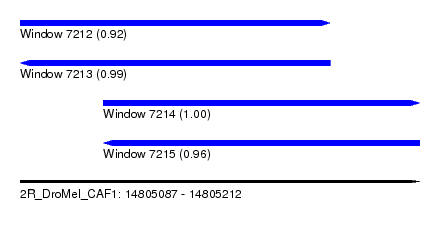
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,805,087 – 14,805,212 |
| Length | 125 |
| Max. P | 0.996498 |

| Location | 14,805,087 – 14,805,184 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 89.35 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -22.50 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14805087 97 + 20766785 GGUGAGUAUCCAAUGAUUGGGUUAGAAUCCGCAAGAAAUGCCGUAAACUACCGGCCUUUCUAACCUAUUUAGUGCAUUUUCGCAAUGCACAAUUCGG ..(((((......(((.((((((((((..(....)....((((........))))..)))))))))).)))(((((((.....))))))).))))). ( -30.30) >DroSec_CAF1 6983 97 + 1 GGUGAGUAUCCAAAGAUUGCGUUAGAAUCCGCAAGAAAUGCCGUGAAGUACCGGCAUUUCUAACCUAUUUAGUGCAUUUUCGCAAUGCACAAUUCUG (((..............((((........))))((((((((((........)))))))))).)))......(((((((.....)))))))....... ( -29.10) >DroSim_CAF1 1068 97 + 1 GGUGAGUAUCCAAUGAUUGGGUUAGAAUCCGCAAGAAAUGCCGUGAUCUGCCGGCAUUUCUAACCUAUUUAGUGCAUUUUCGCAAUGCACAAUUCGG ..(((((......(((.(((((((((...(....).(((((((........)))))))))))))))).)))(((((((.....))))))).))))). ( -32.80) >DroYak_CAF1 1067 97 + 1 GGUGAGUACUUAAUGAUUGCAUUAAAACCCGCAGGAAAUGCAGUAGACUACCGGCAUUUCUAACCUAUGAAGUGCAUUUUCGCGAUGCACAAUUCGG .(((.((..((((((....)))))).)).))).((((((((.(((...)))..))))))))......(((((((((((.....)))))))..)))). ( -23.60) >consensus GGUGAGUAUCCAAUGAUUGCGUUAGAAUCCGCAAGAAAUGCCGUAAACUACCGGCAUUUCUAACCUAUUUAGUGCAUUUUCGCAAUGCACAAUUCGG (((....(((((.....)))))....)))....((((((((((........))))))))))..........(((((((.....)))))))....... (-22.50 = -23.00 + 0.50)


| Location | 14,805,087 – 14,805,184 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 89.35 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -25.12 |
| Energy contribution | -25.75 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14805087 97 - 20766785 CCGAAUUGUGCAUUGCGAAAAUGCACUAAAUAGGUUAGAAAGGCCGGUAGUUUACGGCAUUUCUUGCGGAUUCUAACCCAAUCAUUGGAUACUCACC ((((...(((((((.....)))))))......((((((((..((((........))))...((.....))))))))))......))))......... ( -28.50) >DroSec_CAF1 6983 97 - 1 CAGAAUUGUGCAUUGCGAAAAUGCACUAAAUAGGUUAGAAAUGCCGGUACUUCACGGCAUUUCUUGCGGAUUCUAACGCAAUCUUUGGAUACUCACC .(((((((((((((.....))))))).......((.((((((((((........)))))))))).)).))))))......(((....)))....... ( -31.20) >DroSim_CAF1 1068 97 - 1 CCGAAUUGUGCAUUGCGAAAAUGCACUAAAUAGGUUAGAAAUGCCGGCAGAUCACGGCAUUUCUUGCGGAUUCUAACCCAAUCAUUGGAUACUCACC ..((((((((((((.....))))))).......((.((((((((((........)))))))))).)).)))))....(((.....)))......... ( -31.40) >DroYak_CAF1 1067 97 - 1 CCGAAUUGUGCAUCGCGAAAAUGCACUUCAUAGGUUAGAAAUGCCGGUAGUCUACUGCAUUUCCUGCGGGUUUUAAUGCAAUCAUUAAGUACUCACC ..(((..((((((.......)))))))))....((..(((((((.((((...)))))))))))..))(((((((((((....))))))).))))... ( -25.20) >consensus CCGAAUUGUGCAUUGCGAAAAUGCACUAAAUAGGUUAGAAAUGCCGGUAGUUCACGGCAUUUCUUGCGGAUUCUAACCCAAUCAUUGGAUACUCACC ..((((((((((((.....))))))).......((.((((((((((........)))))))))).)).)))))........................ (-25.12 = -25.75 + 0.62)

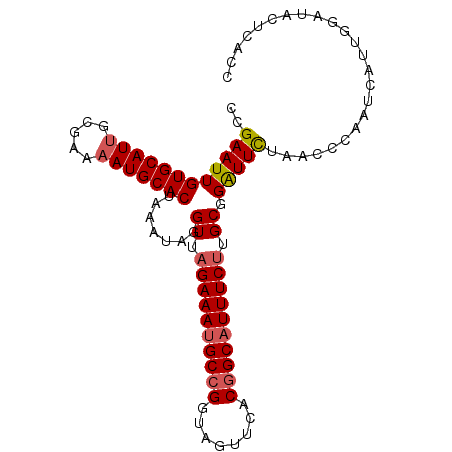
| Location | 14,805,113 – 14,805,212 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 90.92 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -27.35 |
| Energy contribution | -27.97 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14805113 99 + 20766785 AUCCGCAAGAAAUGCCGUAAACUACCGGCCUUUCUAACCUAUUUAGUGCAUUUUCGCAAUGCACAAUUCGGA--AAUACCUGCGACAAUAUGGUCGUCCCG .((((..(((((.((((........)))).)))))..........(((((((.....)))))))....))))--.......(((((......))))).... ( -28.30) >DroSec_CAF1 7009 101 + 1 AUCCGCAAGAAAUGCCGUGAAGUACCGGCAUUUCUAACCUAUUUAGUGCAUUUUCGCAAUGCACAAUUCUGAAAAAUACCUGCGACAAUAUGGUCGUCCCG ....(((((((((((((........))))))))))..........(((((((.....)))))))................)))(((......)))...... ( -28.60) >DroSim_CAF1 1094 101 + 1 AUCCGCAAGAAAUGCCGUGAUCUGCCGGCAUUUCUAACCUAUUUAGUGCAUUUUCGCAAUGCACAAUUCGGAAAAAUACCUGCGACAAUAUGGUCGUCCCG .((((..((((((((((........))))))))))..........(((((((.....)))))))....)))).........(((((......))))).... ( -32.60) >DroYak_CAF1 1093 101 + 1 ACCCGCAGGAAAUGCAGUAGACUACCGGCAUUUCUAACCUAUGAAGUGCAUUUUCGCGAUGCACAAUUCGGAAAAUUACCUGCGACAUUAUGGUCGUCUCG ....(((((..................(((((((........)))))))((((((.(((........)))))))))..)))))(((......)))...... ( -27.30) >consensus AUCCGCAAGAAAUGCCGUAAACUACCGGCAUUUCUAACCUAUUUAGUGCAUUUUCGCAAUGCACAAUUCGGAAAAAUACCUGCGACAAUAUGGUCGUCCCG .((((..((((((((((........))))))))))..........(((((((.....)))))))....)))).........(((((......))))).... (-27.35 = -27.97 + 0.62)


| Location | 14,805,113 – 14,805,212 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 90.92 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -28.49 |
| Energy contribution | -29.80 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14805113 99 - 20766785 CGGGACGACCAUAUUGUCGCAGGUAUU--UCCGAAUUGUGCAUUGCGAAAAUGCACUAAAUAGGUUAGAAAGGCCGGUAGUUUACGGCAUUUCUUGCGGAU .(.(((((.....))))).).......--(((.....(((((((.....))))))).......((.(((((.((((........)))).))))).))))). ( -31.30) >DroSec_CAF1 7009 101 - 1 CGGGACGACCAUAUUGUCGCAGGUAUUUUUCAGAAUUGUGCAUUGCGAAAAUGCACUAAAUAGGUUAGAAAUGCCGGUACUUCACGGCAUUUCUUGCGGAU .(.(((((.....))))).)........(((......(((((((.....))))))).......((.((((((((((........)))))))))).))))). ( -32.70) >DroSim_CAF1 1094 101 - 1 CGGGACGACCAUAUUGUCGCAGGUAUUUUUCCGAAUUGUGCAUUGCGAAAAUGCACUAAAUAGGUUAGAAAUGCCGGCAGAUCACGGCAUUUCUUGCGGAU .(.(((((.....))))).).........(((.....(((((((.....))))))).......((.((((((((((........)))))))))).))))). ( -35.60) >DroYak_CAF1 1093 101 - 1 CGAGACGACCAUAAUGUCGCAGGUAAUUUUCCGAAUUGUGCAUCGCGAAAAUGCACUUCAUAGGUUAGAAAUGCCGGUAGUCUACUGCAUUUCCUGCGGGU ..(((((((......)))((.((((.((((((.....((((((.......))))))......))..)))).)))).)).)))).(((((.....))))).. ( -26.50) >consensus CGGGACGACCAUAUUGUCGCAGGUAUUUUUCCGAAUUGUGCAUUGCGAAAAUGCACUAAAUAGGUUAGAAAUGCCGGUAGUUCACGGCAUUUCUUGCGGAU .(.(((((.....))))).).........(((.....(((((((.....))))))).......((.((((((((((........)))))))))).))))). (-28.49 = -29.80 + 1.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:34 2006