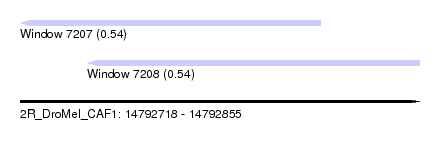
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,792,718 – 14,792,855 |
| Length | 137 |
| Max. P | 0.540793 |

| Location | 14,792,718 – 14,792,821 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -28.43 |
| Energy contribution | -29.93 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
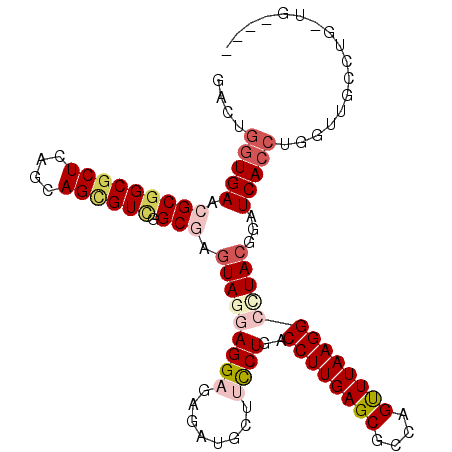
>2R_DroMel_CAF1 14792718 103 - 20766785 GACUGGUGAAGGCGGCGCUCAGCAGCGUCGGCGAGUAGAAGGAUAGAUGCUUCCUGACCUUGAGCGCCAGUUUAAGGCCUACGGAUCACCUGGUUGCCUGGUU---- (((((((((.(.(((((((....))))))).)..((((.((((........))))..((((((((....)))))))).))))...))))).))))........---- ( -39.50) >DroVir_CAF1 26235 106 - 1 GGCUGGUGAACGCGGCGCUCAGCAGUGUGGGCGAGUAUGAGGCGAGGUGCUUUCUGACCUUGAGCGCCAGCUUAAGGCUUACGGUUCACCAG-UUGCCUGUUGUUGU (((((((((((..((((((((..((.((.((.((((((........)))))).)).))))))))))))(((.....)))....)))))))))-))............ ( -44.70) >DroGri_CAF1 28642 98 - 1 GUGAGGUGAACGCGGCGCUCAGCAGCGUCGGCGAAUAUGAGGCGAGAUGCUUCCUGACCUUGAGCGCCAGCUUAAGGCUUACACUUCACCAC-UUGGCC-------- ((((((((..(((((((((....)))))).))).....(((((.....)))))....((((((((....))))))))....))))))))...-......-------- ( -39.90) >DroEre_CAF1 26811 103 - 1 GACUGGUGAAGGCGGCGCUCAGCAGCGUCGGCGAGUAGAAGGAUAGAUGCUUCCUGACCUUGAGCGCCAGUUUAAGGCCUACGGAUCACCUGGUUGCCUGGCC---- ....(((..((((((((((((..((.(((((.(((((..........))))).))))))))))))))).......((((...((....)).)))))))).)))---- ( -40.40) >DroWil_CAF1 36957 98 - 1 AACUUGUGAACGCGGCACUCAGCAGCGUUGGCGAGUAGGAGGAUAGAUGUUUCCUGACCUUGAGCGCCAGUUUAAGGCUUACACUUCAACGGGUUUC-----G---- ...((((.(((((.((.....)).))))).))))(((((((((........))))..((((((((....)))))))))))))...............-----.---- ( -31.70) >DroAna_CAF1 22520 103 - 1 GGCUGGUGAACGCGGCGCUGAGCAGCGUCGGCGAGUAGGAGGAGAGGUGCUUCCUGACCUUGAGCGCCAGUUUAAGGCCUACGGAUCACCUGGUUGCCUGUUG---- (((.(((((.(((((((((....)))))).))).((((((((((......)))))..((((((((....)))))))))))))...))))).....))).....---- ( -45.60) >consensus GACUGGUGAACGCGGCGCUCAGCAGCGUCGGCGAGUAGGAGGAGAGAUGCUUCCUGACCUUGAGCGCCAGUUUAAGGCCUACGGAUCACCUGGUUGCCUG_UG____ ....(((((.(((((((((....)))))).))).(((((((((........))))..((((((((....)))))))))))))...)))))................. (-28.43 = -29.93 + 1.50)

| Location | 14,792,741 – 14,792,855 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.08 |
| Mean single sequence MFE | -43.77 |
| Consensus MFE | -29.51 |
| Energy contribution | -29.35 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
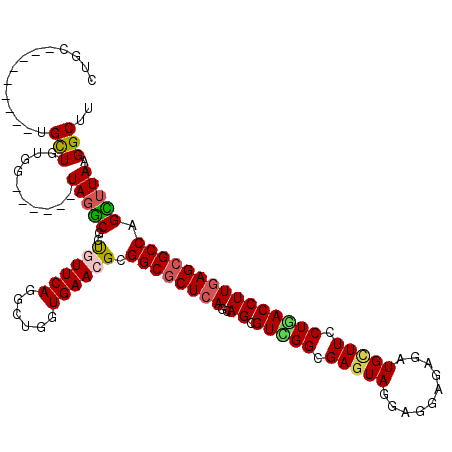
>2R_DroMel_CAF1 14792741 114 - 20766785 CUGCUAUUGCUGCUGUUGUUG------UACACACCGUUCAGACUGGUGAAGGCGGCGCUCAGCAGCGUCGGCGAGUAGAAGGAUAGAUGCUUCCUGACCUUGAGCGCCAGUUUAAGGCCU .((((..((((((((.(((((------(...(((((.......)))))...))))))..))))))))..))))......((((........))))..((((((((....))))))))... ( -43.20) >DroVir_CAF1 26261 105 - 1 CUGC---------UGCUGUGA------UAGGCGCUGUUCAGGCUGGUGAACGCGGCGCUCAGCAGUGUGGGCGAGUAUGAGGCGAGGUGCUUUCUGACCUUGAGCGCCAGCUUAAGGCUU ..((---------(((((.(.------..(.(((.(((((......)))))))).).).)))))))......((((...((((..(((((((.........))))))).))))...)))) ( -41.40) >DroGri_CAF1 28660 105 - 1 CUGC---------UGCUGUGG------UAGGCGCUGUUCAGUGAGGUGAACGCGGCGCUCAGCAGCGUCGGCGAAUAUGAGGCGAGAUGCUUCCUGACCUUGAGCGCCAGCUUAAGGCUU ..((---------(((((.((------(.(.(((.(((((......)))))))).)))))))))))(((((.......(((((.....))))))))))(((((((....))))))).... ( -43.31) >DroWil_CAF1 36975 105 - 1 CUGC---------UGCUGUGA------UAGACGCUAUUCAAACUUGUGAACGCGGCACUCAGCAGCGUUGGCGAGUAGGAGGAUAGAUGUUUCCUGACCUUGAGCGCCAGUUUAAGGCUU ....---------........------.(((((((((((..((((((.(((((.((.....)).))))).))))))....)))))).))))).....((((((((....))))))))... ( -34.50) >DroAna_CAF1 22543 120 - 1 CUGCUGCUGUUGCUGUUGCUGUGGCGGUAGGCACCGUUCAGGCUGGUGAACGCGGCGCUGAGCAGCGUCGGCGAGUAGGAGGAGAGGUGCUUCCUGACCUUGAGCGCCAGUUUAAGGCCU (((((((..(.((....)).)..))))))).........(((((..(((((..((((((....))))))((((..(((((((((......)))))...))))..)))).))))).))))) ( -51.70) >DroMoj_CAF1 30415 105 - 1 CUGC---------UGCUGUGG------UAGGCGCUGUUCAGGCUGGUGAACGCGGCGCUCAGCAGUGUGGGGGAGUACGAGGCAAGAUGCUUCCUCACCUUGAGCGCCAGCUUAAGGCUU ....---------.(((....------(((((((.(((((......)))))))((((((((..((.((((((((((((.......).))))))))))))))))))))).))))).))).. ( -48.50) >consensus CUGC_________UGCUGUGG______UAGGCGCUGUUCAGGCUGGUGAACGCGGCGCUCAGCAGCGUCGGCGAGUAGGAGGAGAGAUGCUUCCUGACCUUGAGCGCCAGCUUAAGGCUU ..............(((..........(((((..((((((......)))))).((((((((..((.(((((.(((((..........))))).))))))))))))))).))))).))).. (-29.51 = -29.35 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:27 2006