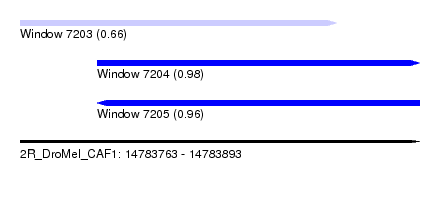
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,783,763 – 14,783,893 |
| Length | 130 |
| Max. P | 0.982696 |

| Location | 14,783,763 – 14,783,866 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.36 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -22.12 |
| Energy contribution | -23.60 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
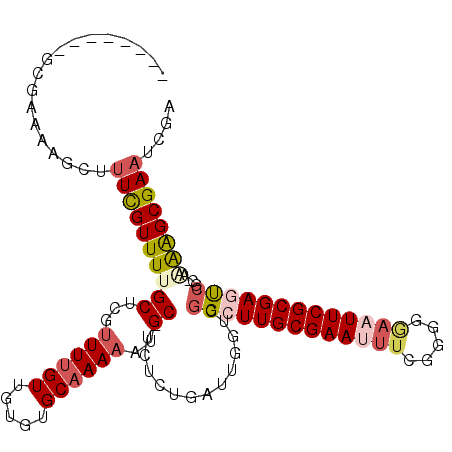
>2R_DroMel_CAF1 14783763 103 + 20766785 --------GCGAAAAGCUUUCGUUUUGCUCUUUUUGUUGUGUGCAAAAAUGC-UCUCUGAUUGGUCGGCUUGCGAAUUUCGGGGGAAUUCGCGAGUCGC-AAAAGCGAAUCGA --------((.....)).((((((((((..(((((((.....))))))).))-..........(.((((((((((((((.....)))))))))))))))-.)))))))).... ( -34.50) >DroSec_CAF1 18643 103 + 1 --------GCGAAAAGCUUUUGUUUUGCUCGUUUUGUUGUGUGCAAAAAUGC-UCUCUGAUUGGUCGGCUUGCGAAUUUCGGGGGAAUUCGCGAGUCGC-AAAAGCGAAUCGA --------.(((...((((((((...((...((((((.....))))))..))-.............(((((((((((((.....)))))))))))))))-))))))...))). ( -37.80) >DroEre_CAF1 17839 103 + 1 --------GCGAAAAGCUUUCGUUUUGCUCGUUUUGUUGUGUGCAAAAAUGC-UCUCUGAUUGGUCGGCUUGCGAAUUUCGGAGGAAUUCGCGAGUCGC-AGAAGCGAAUCGA --------((.....)).((((((((((...((((((.....))))))..))-..........(.((((((((((((((.....)))))))))))))))-.)))))))).... ( -35.30) >DroWil_CAF1 25763 113 + 1 AGCUUCAUGGUCAAAGCUUUCGUUUUGCUCUUUUUGUUAUUUGCCAAUUUGCCUCUCUGAUUGGUCGGAUUGCGAAUUUCGUGUAUUUUCGCGAAUGCCGAUGAGCGAAUCCA .(((.((((((((((((....)))))).....(((((.(((((((((((.........)))))).))))).))))).((((((......)))))).))).))))))....... ( -29.10) >DroAna_CAF1 14624 103 + 1 --------GAGAAAAGCUCUUGUUUUGCUCGUUUUGUUGUGUGCAAAAAUGC-UCUCUGAUUGGUCGGAUUGCGAAUUUCGGAGCAUUUCGCGAACCUC-UCAGGCGAAUGGA --------(((.(((((....))))).))).((((((.....))))))((((-(((..((((.((......)).))))..)))))))(((((.......-....))))).... ( -26.40) >consensus ________GCGAAAAGCUUUCGUUUUGCUCGUUUUGUUGUGUGCAAAAAUGC_UCUCUGAUUGGUCGGCUUGCGAAUUUCGGGGGAAUUCGCGAGUCGC_AAAAGCGAAUCGA ..................((((((((((...((((((.....))))))..))..............(((((((((((((.....)))))))))))))....)))))))).... (-22.12 = -23.60 + 1.48)

| Location | 14,783,788 – 14,783,893 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -37.94 |
| Consensus MFE | -21.86 |
| Energy contribution | -24.06 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14783788 105 + 20766785 UUGUUGUGUGCAAAAAUGC-UCUCUGAUUGGUCGGCUUGCGAAUUUCGGGGGAAUUCGCGAGUCGC-AAAAGCGAAUCGAAAGCUUGCACAA------UUUGCCGAGCGG-----CAA ......(((((((....((-(.((.((((.((((((((((((((((.....)))))))))))))).-....)).)))))).)))))))))).------.(((((....))-----))) ( -42.90) >DroSec_CAF1 18668 105 + 1 UUGUUGUGUGCAAAAAUGC-UCUCUGAUUGGUCGGCUUGCGAAUUUCGGGGGAAUUCGCGAGUCGC-AAAAGCGAAUCGAAAGCUUGCACAA------UUUGCCGAGCGG-----CAA ......(((((((....((-(.((.((((.((((((((((((((((.....)))))))))))))).-....)).)))))).)))))))))).------.(((((....))-----))) ( -42.90) >DroEre_CAF1 17864 105 + 1 UUGUUGUGUGCAAAAAUGC-UCUCUGAUUGGUCGGCUUGCGAAUUUCGGAGGAAUUCGCGAGUCGC-AGAAGCGAAUCGAAAGCUUGCACAA------UCUGCCGAGCGG-----CAG ......(((((((....((-(.((.((((.((((((((((((((((.....)))))))))))))).-....)).)))))).)))))))))).------.(((((....))-----))) ( -44.50) >DroWil_CAF1 25796 118 + 1 UUGUUAUUUGCCAAUUUGCCUCUCUGAUUGGUCGGAUUGCGAAUUUCGUGUAUUUUCGCGAAUGCCGAUGAGCGAAUCCAAAGUUUGCACAUAAGAGCUUCGUCUAGCCGACACACAA ..(..((((((.((((((((.........)).))))))))))))..)((((......(((((.((..(((.((((((.....)))))).)))....))))))).......)))).... ( -29.52) >DroAna_CAF1 14649 104 + 1 UUGUUGUGUGCAAAAAUGC-UCUCUGAUUGGUCGGAUUGCGAAUUUCGGAGCAUUUCGCGAACCUC-UCAGGCGAAUGGAAAGCUUGCAAGA------A-AGCGUCUCGG-----GAG .....((.(((..((((((-(((..((((.((......)).))))..))))))))).))).))(((-((((((...(....)((((......------)-))))))).))-----))) ( -29.90) >consensus UUGUUGUGUGCAAAAAUGC_UCUCUGAUUGGUCGGCUUGCGAAUUUCGGGGGAAUUCGCGAGUCGC_AAAAGCGAAUCGAAAGCUUGCACAA______UUUGCCGAGCGG_____CAA ......(((((((....((.....(((((.((((((((((((((((.....))))))))))))))......)).)))))...)))))))))........................... (-21.86 = -24.06 + 2.20)


| Location | 14,783,788 – 14,783,893 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -31.76 |
| Consensus MFE | -14.60 |
| Energy contribution | -17.04 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14783788 105 - 20766785 UUG-----CCGCUCGGCAAA------UUGUGCAAGCUUUCGAUUCGCUUUU-GCGACUCGCGAAUUCCCCCGAAAUUCGCAAGCCGACCAAUCAGAGA-GCAUUUUUGCACACAACAA (((-----((....))))).------.(((((((((((((((.((((....-)))).))(((((((.......)))))))..............))))-))....)))))))...... ( -37.10) >DroSec_CAF1 18668 105 - 1 UUG-----CCGCUCGGCAAA------UUGUGCAAGCUUUCGAUUCGCUUUU-GCGACUCGCGAAUUCCCCCGAAAUUCGCAAGCCGACCAAUCAGAGA-GCAUUUUUGCACACAACAA (((-----((....))))).------.(((((((((((((((.((((....-)))).))(((((((.......)))))))..............))))-))....)))))))...... ( -37.10) >DroEre_CAF1 17864 105 - 1 CUG-----CCGCUCGGCAGA------UUGUGCAAGCUUUCGAUUCGCUUCU-GCGACUCGCGAAUUCCUCCGAAAUUCGCAAGCCGACCAAUCAGAGA-GCAUUUUUGCACACAACAA (((-----((....))))).------.(((((((((((((((.((((....-)))).))(((((((.......)))))))..............))))-))....)))))))...... ( -39.10) >DroWil_CAF1 25796 118 - 1 UUGUGUGUCGGCUAGACGAAGCUCUUAUGUGCAAACUUUGGAUUCGCUCAUCGGCAUUCGCGAAAAUACACGAAAUUCGCAAUCCGACCAAUCAGAGAGGCAAAUUGGCAAAUAACAA .(((.((((((((......))))......(((...(((((((.....)).((((.(((.(((((...........)))))))))))).....)))))..)))....))))....))). ( -24.70) >DroAna_CAF1 14649 104 - 1 CUC-----CCGAGACGCU-U------UCUUGCAAGCUUUCCAUUCGCCUGA-GAGGUUCGCGAAAUGCUCCGAAAUUCGCAAUCCGACCAAUCAGAGA-GCAUUUUUGCACACAACAA ...-----..((((.(((-(------......))))))))...((..((((-..((((.(((((.(.......).))))).....))))..)))).))-(((....)))......... ( -20.80) >consensus UUG_____CCGCUCGGCAAA______UUGUGCAAGCUUUCGAUUCGCUUUU_GCGACUCGCGAAUUCCCCCGAAAUUCGCAAGCCGACCAAUCAGAGA_GCAUUUUUGCACACAACAA ...........................(((((((((((((((.((((.....)))).))(((((((.......)))))))..............)))).))....)))))))...... (-14.60 = -17.04 + 2.44)

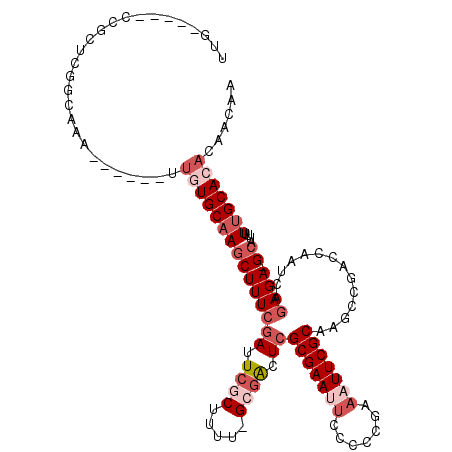
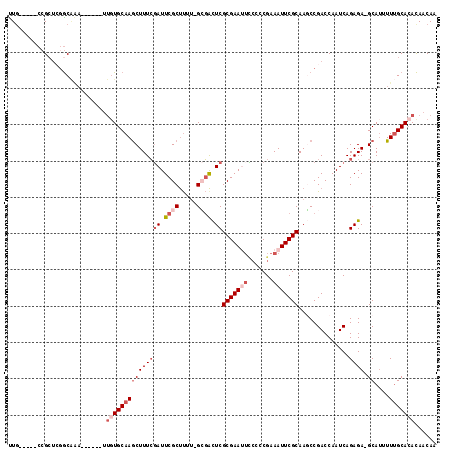
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:24 2006