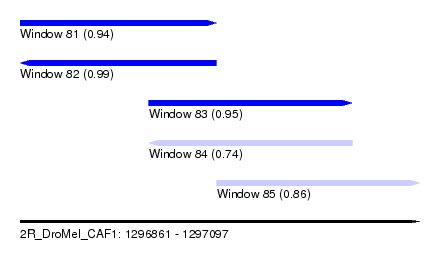
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,296,861 – 1,297,097 |
| Length | 236 |
| Max. P | 0.986227 |

| Location | 1,296,861 – 1,296,977 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 96.84 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -27.75 |
| Energy contribution | -27.19 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1296861 116 + 20766785 UAUUUUGCGUAAAAUACGUCCUACCCUACACAUCAAACGCUGCUCUCUGCGAUGGCGUGCAAUACCCAUGCCACAGUCCUUGAUGUAUAGCUUGGGCAAUAUCAGCAUUUUAUUCU .....((((((...)))((((....(((.(((((((.(((........))).(((((((.......)))))))......))))))).)))...)))).......)))......... ( -30.60) >DroSec_CAF1 24877 116 + 1 UAUUUUGCGUAAAAUACGUCUUACCCCACACAUCAAACGCUGCUCUCUGCGAUGGCGUGCUAUACCCAUGCCACAGCCCUUGAUGUAUAGCUUGGGCAAUAUCAGCAUUUUAUUCU .....(((((((........))))((((.(((((((..(((((.....))(.(((((((.......)))))))))))..)))))))......))))........)))......... ( -27.30) >DroSim_CAF1 29075 116 + 1 UAUUUUGCGUAAAAUACGUCCUACCCCACACAUCAAACGCUGCUCUCUGCGAUGGCGUGCAAUACCCAUGCCACAGCCCUUGAUGUAUAGCUUGGGCAAUAUCAGCAUUUUAUUCU .....((((((...)))((((........(((((((..(((((.....))(.(((((((.......)))))))))))..))))))).......)))).......)))......... ( -28.76) >DroEre_CAF1 33706 116 + 1 UAUUUUGCGUAAAAUACGUUCUACCAUACACAUCAAACGCUGCUCUCUGCGAUGGCGUGCAAUACUCAUGCCACAGCCCUUGAUGUAUAGCUUGGGCAAUAUCAGCAUUUUAUUCU .....((((((...)))((((.....((.(((((((..(((((.....))(.(((((((.......)))))))))))..))))))).))....)))).......)))......... ( -27.00) >consensus UAUUUUGCGUAAAAUACGUCCUACCCCACACAUCAAACGCUGCUCUCUGCGAUGGCGUGCAAUACCCAUGCCACAGCCCUUGAUGUAUAGCUUGGGCAAUAUCAGCAUUUUAUUCU .....((((((...)))((((........(((((((..(((((.....))(.(((((((.......)))))))))))..))))))).......)))).......)))......... (-27.75 = -27.19 + -0.56)

| Location | 1,296,861 – 1,296,977 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 96.84 |
| Mean single sequence MFE | -38.83 |
| Consensus MFE | -37.39 |
| Energy contribution | -37.45 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1296861 116 - 20766785 AGAAUAAAAUGCUGAUAUUGCCCAAGCUAUACAUCAAGGACUGUGGCAUGGGUAUUGCACGCCAUCGCAGAGAGCAGCGUUUGAUGUGUAGGGUAGGACGUAUUUUACGCAAAAUA .(..((((((((.....((((((....(((((((((((..((((.((((((((.....)).)))).)).....))))..)))))))))))))))))...))))))))..)...... ( -36.40) >DroSec_CAF1 24877 116 - 1 AGAAUAAAAUGCUGAUAUUGCCCAAGCUAUACAUCAAGGGCUGUGGCAUGGGUAUAGCACGCCAUCGCAGAGAGCAGCGUUUGAUGUGUGGGGUAAGACGUAUUUUACGCAAAAUA .(..((((((((.....((((((....(((((((((((.(((((.((((((((.....)).)))).)).....))))).)))))))))))))))))...))))))))..)...... ( -42.00) >DroSim_CAF1 29075 116 - 1 AGAAUAAAAUGCUGAUAUUGCCCAAGCUAUACAUCAAGGGCUGUGGCAUGGGUAUUGCACGCCAUCGCAGAGAGCAGCGUUUGAUGUGUGGGGUAGGACGUAUUUUACGCAAAAUA .(..((((((((.....((((((....(((((((((((.(((((.((((((((.....)).)))).)).....))))).)))))))))))))))))...))))))))..)...... ( -41.70) >DroEre_CAF1 33706 116 - 1 AGAAUAAAAUGCUGAUAUUGCCCAAGCUAUACAUCAAGGGCUGUGGCAUGAGUAUUGCACGCCAUCGCAGAGAGCAGCGUUUGAUGUGUAUGGUAGAACGUAUUUUACGCAAAAUA .(..((((((((.....(((((.....(((((((((((.((((((((.((.......)).))).((.....))))))).))))))))))).)))))...))))))))..)...... ( -35.20) >consensus AGAAUAAAAUGCUGAUAUUGCCCAAGCUAUACAUCAAGGGCUGUGGCAUGGGUAUUGCACGCCAUCGCAGAGAGCAGCGUUUGAUGUGUAGGGUAGGACGUAUUUUACGCAAAAUA .(..((((((((.....((((((....(((((((((((.((((((((.((.......)).))).((.....))))))).)))))))))))))))))...))))))))..)...... (-37.39 = -37.45 + 0.06)

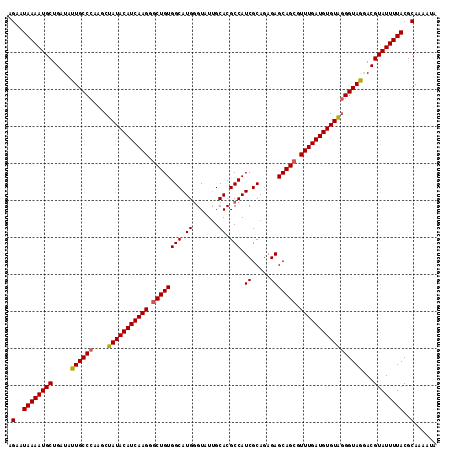
| Location | 1,296,937 – 1,297,057 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -40.25 |
| Consensus MFE | -36.79 |
| Energy contribution | -36.79 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1296937 120 + 20766785 UCCUUGAUGUAUAGCUUGGGCAAUAUCAGCAUUUUAUUCUGCGUUCUCGUCAGUUUCCGGUGCUGCCACUGCUGCUGUUUGAAGCAGAGCCAGCGGUGGCAUCCGCCAUCAUCAUCAUUG ....(((((.........(((......((((((..(..(((((....)).)))..)..))))))((((((((((((((.....))))...))))))))))....))).....)))))... ( -40.94) >DroSec_CAF1 24953 120 + 1 CCCUUGAUGUAUAGCUUGGGCAAUAUCAGCAUUUUAUUCUGCGUUCUCGUCAGUUUCCGGUGCUGCCACUGCUGCUGUUUGAAACAGAGCCAGCGGUGGCAUCCGCCAUCAUCAUCAUUG ....(((((.........(((......((((((..(..(((((....)).)))..)..))))))((((((((((((((.....))))...))))))))))....))).....)))))... ( -40.34) >DroSim_CAF1 29151 120 + 1 CCCUUGAUGUAUAGCUUGGGCAAUAUCAGCAUUUUAUUCUGCGUUCUCGUUAGUUUCCGGUGCUGCCACUACUGCUGUUGGAAGCAGAGCCAGCGGUGGCAUCCGCCAUCAUCAUCAUUG ....(((((....(((..(((.....(((((((..(..(((((....)).)))..)..)))))))......(((((......))))).))))))((((((....))))))..)))))... ( -38.80) >DroEre_CAF1 33782 120 + 1 CCCUUGAUGUAUAGCUUGGGCAAUAUCAGCAUUUUAUUCUGCGUUCUCGUCAGUUUCCGCUGCUGCCACUGCUGUUAUUUGAAGCAGAGGCGGCGGUGGCAUCCGCCAUCAUCAUCAUUG ....(((((.........(((.................(((((....)).)))...(((((((((((.(((((.........))))).))))))))))).....))).....)))))... ( -40.94) >consensus CCCUUGAUGUAUAGCUUGGGCAAUAUCAGCAUUUUAUUCUGCGUUCUCGUCAGUUUCCGGUGCUGCCACUGCUGCUGUUUGAAGCAGAGCCAGCGGUGGCAUCCGCCAUCAUCAUCAUUG ....(((((.........(((......((((((..(..(((((....)).)))..)..))))))((((((((((((((.....))))...))))))))))....))).....)))))... (-36.79 = -36.79 + -0.00)

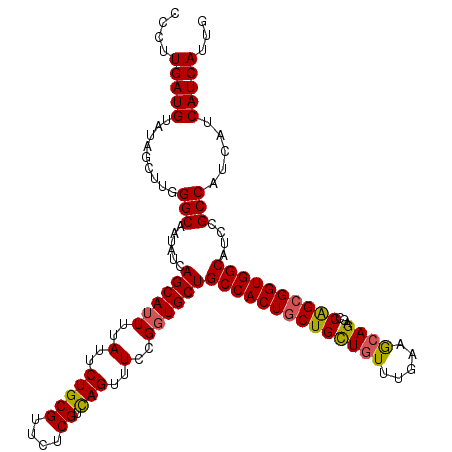
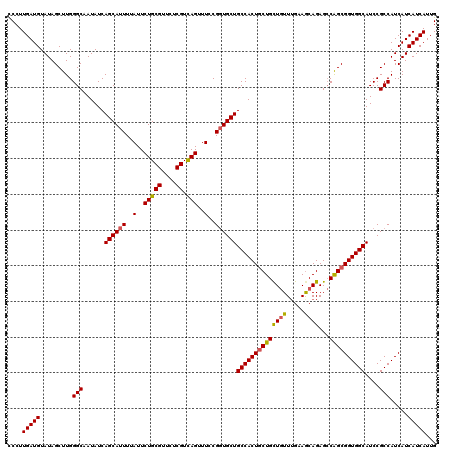
| Location | 1,296,937 – 1,297,057 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -36.85 |
| Consensus MFE | -32.33 |
| Energy contribution | -32.45 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1296937 120 - 20766785 CAAUGAUGAUGAUGGCGGAUGCCACCGCUGGCUCUGCUUCAAACAGCAGCAGUGGCAGCACCGGAAACUGACGAGAACGCAGAAUAAAAUGCUGAUAUUGCCCAAGCUAUACAUCAAGGA ......((((((((((((.(((..((((((...(((((......)))))))))))..)))))((...(((.((....)))))........((.......))))..))))).))))).... ( -38.30) >DroSec_CAF1 24953 120 - 1 CAAUGAUGAUGAUGGCGGAUGCCACCGCUGGCUCUGUUUCAAACAGCAGCAGUGGCAGCACCGGAAACUGACGAGAACGCAGAAUAAAAUGCUGAUAUUGCCCAAGCUAUACAUCAAGGG ......((((((((((((.((((((.((((...((((.....)))))))).))))))...))((...(((.((....)))))........((.......))))..))))).))))).... ( -38.70) >DroSim_CAF1 29151 120 - 1 CAAUGAUGAUGAUGGCGGAUGCCACCGCUGGCUCUGCUUCCAACAGCAGUAGUGGCAGCACCGGAAACUAACGAGAACGCAGAAUAAAAUGCUGAUAUUGCCCAAGCUAUACAUCAAGGG ......((((((((((((.(((..((((((...(((((......)))))))))))..)))))((...(....).....(((........))).........))..))))).))))).... ( -35.00) >DroEre_CAF1 33782 120 - 1 CAAUGAUGAUGAUGGCGGAUGCCACCGCCGCCUCUGCUUCAAAUAACAGCAGUGGCAGCAGCGGAAACUGACGAGAACGCAGAAUAAAAUGCUGAUAUUGCCCAAGCUAUACAUCAAGGG ......(((((..(((((......)))))(((.(((((.........))))).)))..(((((....(((.((....))))).......))))).................))))).... ( -35.40) >consensus CAAUGAUGAUGAUGGCGGAUGCCACCGCUGGCUCUGCUUCAAACAGCAGCAGUGGCAGCACCGGAAACUGACGAGAACGCAGAAUAAAAUGCUGAUAUUGCCCAAGCUAUACAUCAAGGG ......((((((((((((.((((((.((((...(((.......))))))).))))))(((..(....)..........(((........)))......)))))..))))).))))).... (-32.33 = -32.45 + 0.12)

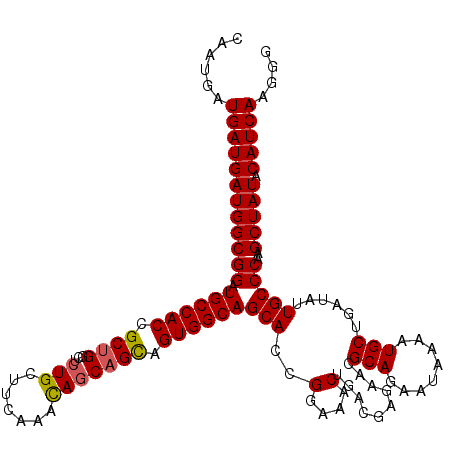
| Location | 1,296,977 – 1,297,097 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -40.66 |
| Consensus MFE | -36.33 |
| Energy contribution | -36.33 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1296977 120 + 20766785 GCGUUCUCGUCAGUUUCCGGUGCUGCCACUGCUGCUGUUUGAAGCAGAGCCAGCGGUGGCAUCCGCCAUCAUCAUCAUUGCUAGCAUUUUGUGCGCACUGACCCGCUUUUUGUGCUUUUC (((.....((((((....((((.(((((((((((((((.....))))...)))))))))))..))))............((..((((...)))))))))))).))).............. ( -41.50) >DroSec_CAF1 24993 120 + 1 GCGUUCUCGUCAGUUUCCGGUGCUGCCACUGCUGCUGUUUGAAACAGAGCCAGCGGUGGCAUCCGCCAUCAUCAUCAUUGCUAGCAUUUUGUGCGCACUGACCCGCUUUUUGUGCUUUUC (((.....((((((....((((.(((((((((((((((.....))))...)))))))))))..))))............((..((((...)))))))))))).))).............. ( -40.90) >DroSim_CAF1 29191 120 + 1 GCGUUCUCGUUAGUUUCCGGUGCUGCCACUACUGCUGUUGGAAGCAGAGCCAGCGGUGGCAUCCGCCAUCAUCAUCAUUGCUAGCAUUUUGUGCGCACUGACCCGCUUUUUGUGCUUUUC (((.....((((((....(((((((...((.(((((......))))))).))))((((((....))))))...)))...))))))......)))((((.............))))..... ( -34.82) >DroEre_CAF1 33822 120 + 1 GCGUUCUCGUCAGUUUCCGCUGCUGCCACUGCUGUUAUUUGAAGCAGAGGCGGCGGUGGCAUCCGCCAUCAUCAUCAUUGCUGGCAUUUUGUGCGCACUGACCCGCUUUUUGUGCUUUUC (((.....((((((...((((((((((.(((((.........))))).))))))))))((((..((((.((.......)).)))).....))))..)))))).))).............. ( -45.40) >consensus GCGUUCUCGUCAGUUUCCGGUGCUGCCACUGCUGCUGUUUGAAGCAGAGCCAGCGGUGGCAUCCGCCAUCAUCAUCAUUGCUAGCAUUUUGUGCGCACUGACCCGCUUUUUGUGCUUUUC (((.....((((((....((((.(((((((((((((((.....))))...)))))))))))..))))............((..((((...)))))))))))).))).............. (-36.33 = -36.33 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:37 2006