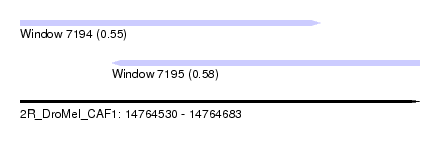
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,764,530 – 14,764,683 |
| Length | 153 |
| Max. P | 0.577735 |

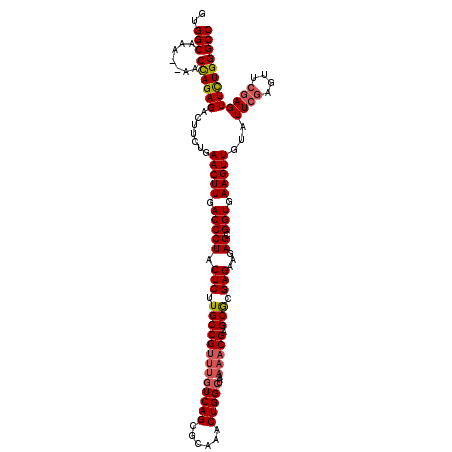
| Location | 14,764,530 – 14,764,645 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.59 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -15.98 |
| Energy contribution | -20.87 |
| Covariance contribution | 4.89 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14764530 115 + 20766785 -----CAGCCAAACAAUUUCAGCUGAGCGAAGCGUAAACUGGCCCAAAACUCGAACUCGAAUACAACUUUACCGCUCUUCUCGCGCUCGUUUUUAGCCAGUUUGCGCUGACAAACGGCAA -----................((((.....(((((((((((((..((((((((....)))............(((.......)))...)))))..)))))))))))))......)))).. ( -36.00) >DroSim_CAF1 5465 95 + 1 -----CAGCCAAACAAUUUCAGCUGAGCGAAGCGUAAACUGGCCCAGAACUCGAACUCGAAUACAACUUCACCGCUCUUCUCGUGCUCGU--------------------CGAACGGCAA -----..(((..............(((((..((........)).......(((....)))............)))))...(((.......--------------------)))..))).. ( -17.20) >DroYak_CAF1 6397 114 + 1 AAAAGCAGCCAAACAAUUUCAGCUGAGCGAAGCGUAAACUGGCCCAGAACUCG------AACACAACUUCACCGCUCUUCUCUCGCUCGUUUUUAGCCAGUUUGCGCUGACAAUCGGCAA .....................(((((....(((((((((((((..(((((..(------((......)))...((.........))..)))))..))))))))))))).....))))).. ( -35.40) >consensus _____CAGCCAAACAAUUUCAGCUGAGCGAAGCGUAAACUGGCCCAGAACUCGAACUCGAAUACAACUUCACCGCUCUUCUCGCGCUCGUUUUUAGCCAGUUUGCGCUGACAAACGGCAA .......(((...........(((......)))((((((((((..((((((((....))).............((.........))..)))))..))))))))))..........))).. (-15.98 = -20.87 + 4.89)

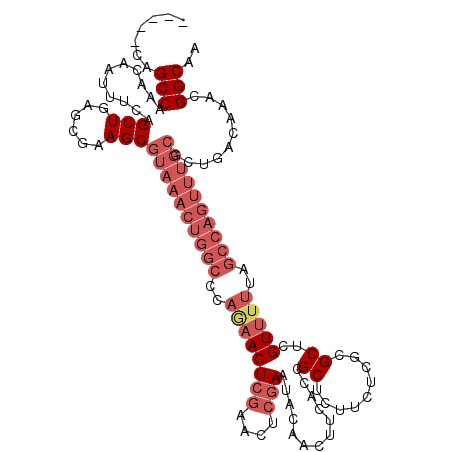
| Location | 14,764,565 – 14,764,683 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -35.31 |
| Consensus MFE | -26.00 |
| Energy contribution | -27.68 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14764565 118 - 20766785 GUGGCAAA--AACCAGAGACUUCUGAACUUGACCCUACUCUUGCCGUUUGUCAGCGCAAACUGGCUAAAAACGAGCGCGAGAAGAGCGGUAAAGUUGUAUUCGAGUUCGAGUUUUGGGCC ..(((...--..((((((.(((..((((((((....(((.(((((((((.((.((((.................))))..)).)))))))))))).....)))))))))))))))))))) ( -37.73) >DroSim_CAF1 5500 98 - 1 GUGGCAAA--AACCAGAGACUUCUGAACUUGACCCUACUCUUGCCGUUCG--------------------ACGAGCACGAGAAGAGCGGUGAAGUUGUAUUCGAGUUCGAGUUCUGGGCC ..(((...--..((((((.(((..((((((((....(((.(..((((((.--------------------.(........)..))))))..)))).....)))))))))))))))))))) ( -34.40) >DroYak_CAF1 6437 114 - 1 GUGGCAAAAAAACUAGAGACUUCUGAACUUGACCCUACUCUUGCCGAUUGUCAGCGCAAACUGGCUAAAAACGAGCGAGAGAAGAGCGGUGAAGUUGUGUU------CGAGUUCUGGGCC ..(((.......((((((.((((..(((((.(((((.(((((((((...(((((......)))))......)).)))))))...)).))).)))))..)..------.)))))))))))) ( -33.81) >consensus GUGGCAAA__AACCAGAGACUUCUGAACUUGACCCUACUCUUGCCGUUUGUCAGCGCAAACUGGCUAAAAACGAGCGCGAGAAGAGCGGUGAAGUUGUAUUCGAGUUCGAGUUCUGGGCC ..(((.......((((((.......(((((.(((((.(((.(((((((((((((......)))))...))))).))).)))...)).))).)))))...((((....))))))))))))) (-26.00 = -27.68 + 1.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:15 2006