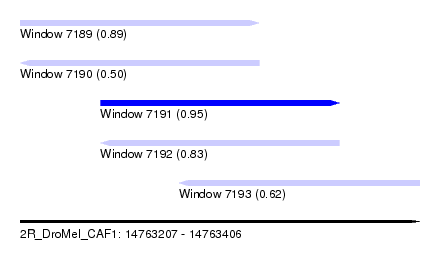
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,763,207 – 14,763,406 |
| Length | 199 |
| Max. P | 0.949916 |

| Location | 14,763,207 – 14,763,326 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.43 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -29.69 |
| Energy contribution | -30.13 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14763207 119 + 20766785 GGUCAGCAUUACAAAUGGAGCACCUCCCAACCAUCCAUCCAAUCCGGUAAUUUUGGGCCU-UUGUGCGAUUGACACACGCAUGUCAGGCCCCAUUACUGGAUCUCCAUCAGGCAUUACCU (((..((.......((((((.....................((((((((((...((((((-.((((((.........))))))..)))))).))))))))))))))))...))...))). ( -36.09) >DroSim_CAF1 4157 119 + 1 GGUCAGCAUUACAAAUAGAGCACCUCCCAACCAUCCAUCCAAUCCGGUAAUUUCGGGCCU-UUGUGCGAUUGACACACGCAUGUCAGGCCCCAUUACUGGAUCUCCAUCAGGCAUUACCU (((..((..........(((...)))...............((((((((((...((((((-.((((((.........))))))..)))))).)))))))))).........))...))). ( -32.20) >DroYak_CAF1 5015 117 + 1 GGUCAGCAGUACAAAUAGAGCACCUACCAACC---GAUCCAAUCCGGUAAUUUCGGGCCUUUUGUGCGAUUGACACACGCAUGUCAGGCCCCAUUACUGAAUCUCCAUCAGGCAUUACCU ((....(((((..................(((---(........))))......((((((..((((((.........))))))..))))))...))))).....))...(((.....))) ( -30.70) >consensus GGUCAGCAUUACAAAUAGAGCACCUCCCAACCAUCCAUCCAAUCCGGUAAUUUCGGGCCU_UUGUGCGAUUGACACACGCAUGUCAGGCCCCAUUACUGGAUCUCCAUCAGGCAUUACCU .(((.((.(((....))).))....................((((((((((...((((((...(((((.........)))))...)))))).))))))))))........)))....... (-29.69 = -30.13 + 0.45)

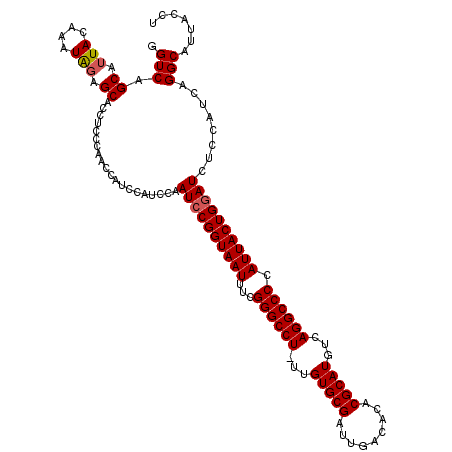
| Location | 14,763,207 – 14,763,326 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.43 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -36.94 |
| Energy contribution | -36.50 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.94 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14763207 119 - 20766785 AGGUAAUGCCUGAUGGAGAUCCAGUAAUGGGGCCUGACAUGCGUGUGUCAAUCGCACAA-AGGCCCAAAAUUACCGGAUUGGAUGGAUGGUUGGGAGGUGCUCCAUUUGUAAUGCUGACC .((((.(((..(((((((..((..((((.((((((....((((.........))))...-))))))...))))((.(((((......))))).)).))..))))))).))).)))).... ( -39.80) >DroSim_CAF1 4157 119 - 1 AGGUAAUGCCUGAUGGAGAUCCAGUAAUGGGGCCUGACAUGCGUGUGUCAAUCGCACAA-AGGCCCGAAAUUACCGGAUUGGAUGGAUGGUUGGGAGGUGCUCUAUUUGUAAUGCUGACC .((((.(((..(((((((..((.......((((((....((((.........))))...-)))))).......((.(((((......))))).)).))..))))))).))).)))).... ( -36.90) >DroYak_CAF1 5015 117 - 1 AGGUAAUGCCUGAUGGAGAUUCAGUAAUGGGGCCUGACAUGCGUGUGUCAAUCGCACAAAAGGCCCGAAAUUACCGGAUUGGAUC---GGUUGGUAGGUGCUCUAUUUGUACUGCUGACC (((.....)))(((...(((((.(((((.((((((....((((.........))))....))))))...))))).)))))..)))---(((..((((.(((.......)))))))..))) ( -41.50) >consensus AGGUAAUGCCUGAUGGAGAUCCAGUAAUGGGGCCUGACAUGCGUGUGUCAAUCGCACAA_AGGCCCGAAAUUACCGGAUUGGAUGGAUGGUUGGGAGGUGCUCUAUUUGUAAUGCUGACC .((((.(((..(((((((((((.(((((.((((((....((((.........))))....))))))...))))).))))(.(((.....))).)......))))))).))).)))).... (-36.94 = -36.50 + -0.44)

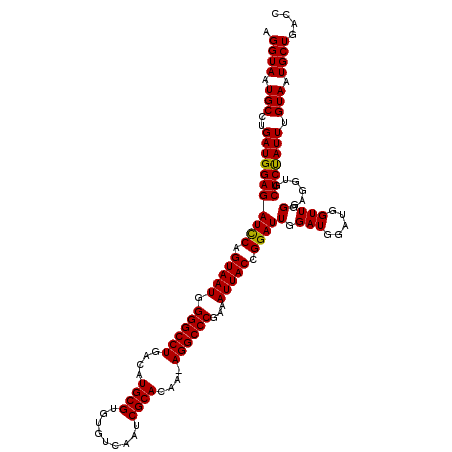
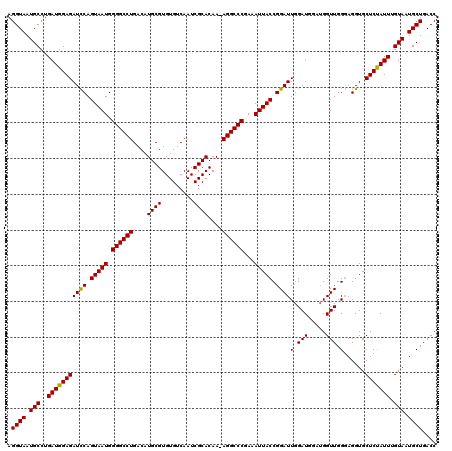
| Location | 14,763,247 – 14,763,366 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -36.25 |
| Consensus MFE | -30.69 |
| Energy contribution | -31.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14763247 119 + 20766785 AAUCCGGUAAUUUUGGGCCU-UUGUGCGAUUGACACACGCAUGUCAGGCCCCAUUACUGGAUCUCCAUCAGGCAUUACCUCCUCCUUACCCUACAUCGCCAGUUAAUUCCAUUGGAAAUU .((((((((((...((((((-.((((((.........))))))..)))))).)))))))))).((((...((.((((.((....................)).)))).))..)))).... ( -33.45) >DroSim_CAF1 4197 115 + 1 AAUCCGGUAAUUUCGGGCCU-UUGUGCGAUUGACACACGCAUGUCAGGCCCCAUUACUGGAUCUCCAUCAGGCAUUACCUCCG----GCCCCGCUUCGCCAGUUAAUUCCUUCGGAAAUU .((((((((((...((((((-.((((((.........))))))..)))))).)))))))))).(((...(((.((((.((..(----((........))))).)))).)))..))).... ( -38.60) >DroYak_CAF1 5052 116 + 1 AAUCCGGUAAUUUCGGGCCUUUUGUGCGAUUGACACACGCAUGUCAGGCCCCAUUACUGAAUCUCCAUCAGGCAUUACCUCCG----GCCCCACAUCGCCAGUUAAUUCCUUUGGAAAUU ....(((((((...((((((..((((((.........))))))..)))))).)))))))....((((..(((.((((.((..(----((........))))).)))).))).)))).... ( -36.70) >consensus AAUCCGGUAAUUUCGGGCCU_UUGUGCGAUUGACACACGCAUGUCAGGCCCCAUUACUGGAUCUCCAUCAGGCAUUACCUCCG____GCCCCACAUCGCCAGUUAAUUCCUUUGGAAAUU .((((((((((...((((((...(((((.........)))))...)))))).))))))))))........(((........................)))......((((...))))... (-30.69 = -31.03 + 0.33)

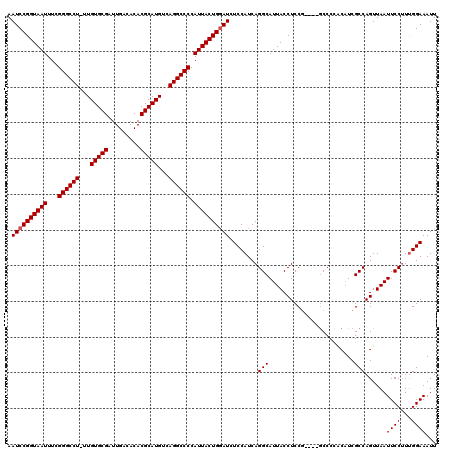
| Location | 14,763,247 – 14,763,366 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -39.47 |
| Consensus MFE | -33.86 |
| Energy contribution | -33.20 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14763247 119 - 20766785 AAUUUCCAAUGGAAUUAACUGGCGAUGUAGGGUAAGGAGGAGGUAAUGCCUGAUGGAGAUCCAGUAAUGGGGCCUGACAUGCGUGUGUCAAUCGCACAA-AGGCCCAAAAUUACCGGAUU ..((((((..........((.((........)).))....(((.....)))..))))))(((.(((((.((((((....((((.........))))...-))))))...))))).))).. ( -34.80) >DroSim_CAF1 4197 115 - 1 AAUUUCCGAAGGAAUUAACUGGCGAAGCGGGGC----CGGAGGUAAUGCCUGAUGGAGAUCCAGUAAUGGGGCCUGACAUGCGUGUGUCAAUCGCACAA-AGGCCCGAAAUUACCGGAUU ..((((((.(((.((((.(((((........))----)))...)))).)))..))))))(((.(((((.((((((....((((.........))))...-))))))...))))).))).. ( -42.70) >DroYak_CAF1 5052 116 - 1 AAUUUCCAAAGGAAUUAACUGGCGAUGUGGGGC----CGGAGGUAAUGCCUGAUGGAGAUUCAGUAAUGGGGCCUGACAUGCGUGUGUCAAUCGCACAAAAGGCCCGAAAUUACCGGAUU ..((((((.(((.((((.(((((........))----)))...)))).)))..))))))(((.(((((.((((((....((((.........))))....))))))...))))).))).. ( -40.90) >consensus AAUUUCCAAAGGAAUUAACUGGCGAUGUGGGGC____CGGAGGUAAUGCCUGAUGGAGAUCCAGUAAUGGGGCCUGACAUGCGUGUGUCAAUCGCACAA_AGGCCCGAAAUUACCGGAUU ..((((((..........((.((...)).)).........(((.....)))..))))))(((.(((((.((((((....((((.........))))....))))))...))))).))).. (-33.86 = -33.20 + -0.66)


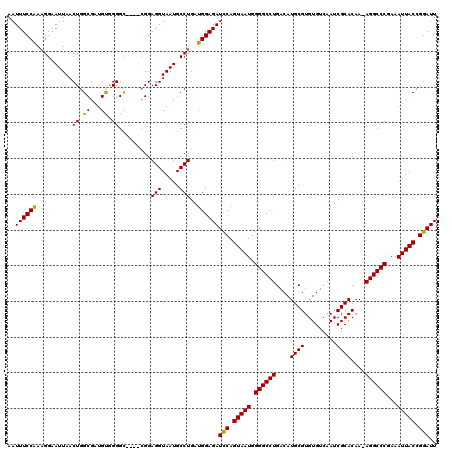
| Location | 14,763,286 – 14,763,406 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.13 |
| Mean single sequence MFE | -38.07 |
| Consensus MFE | -32.67 |
| Energy contribution | -32.23 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14763286 120 - 20766785 UGGUGCGUCAGGUGCAAGGAAAAUCCAAACGGCAAUGCGCAAUUUCCAAUGGAAUUAACUGGCGAUGUAGGGUAAGGAGGAGGUAAUGCCUGAUGGAGAUCCAGUAAUGGGGCCUGACAU ......(((((((..........((((...((((.(((..((((((....))))))..((.((........)).))......))).))))...))))..((((....))))))))))).. ( -32.60) >DroSim_CAF1 4236 116 - 1 UGGUGCGUCAGGUGCAAGGAAAAUCCAAACGGCAAUGCGCAAUUUCCGAAGGAAUUAACUGGCGAAGCGGGGC----CGGAGGUAAUGCCUGAUGGAGAUCCAGUAAUGGGGCCUGACAU ......(((((((..........((((...((((.(((..((((((....))))))..(((((........))----)))..))).))))...))))..((((....))))))))))).. ( -39.90) >DroYak_CAF1 5092 116 - 1 UGGUGCGUCAGGUGCAAGGAAAAUCCAACUGGCAAUGCGCAAUUUCCAAAGGAAUUAACUGGCGAUGUGGGGC----CGGAGGUAAUGCCUGAUGGAGAUUCAGUAAUGGGGCCUGACAU ......(((((((.(..((.....)).((((((.....))((((((((.(((.((((.(((((........))----)))...)))).)))..))))))))))))....).))))))).. ( -41.70) >consensus UGGUGCGUCAGGUGCAAGGAAAAUCCAAACGGCAAUGCGCAAUUUCCAAAGGAAUUAACUGGCGAUGUGGGGC____CGGAGGUAAUGCCUGAUGGAGAUCCAGUAAUGGGGCCUGACAU ......(((((((..........((((...((((.(((..((((((....))))))..((.((...)).))...........))).))))...))))..((((....))))))))))).. (-32.67 = -32.23 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:13 2006