
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,762,106 – 14,762,299 |
| Length | 193 |
| Max. P | 0.994492 |

| Location | 14,762,106 – 14,762,219 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.09 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14762106 113 + 20766785 CC--CCUCCG-CAGACCA----AACUCCAUCUAUUUUCUAUCCAAAUUGUCAGUUGGCCAGUUGGCUCCAACAUUGCAUGUCUUGGCAUACGUGGGCCAUGAAAAAUUCAUGGCCCGCCU ..--.....(-(((((..----..........................))).(((((((....)))..))))..)))((((....))))..(((((((((((.....))))))))))).. ( -33.29) >DroSim_CAF1 2534 114 + 1 CC--CCUCCCCCUCAACA----AACUCCAUCUAUUUUCUAUCCAAAUUGUCAGUUGGCCAGUUGGCUCCAACAUUGCAUGUCUUGGCAUACGUGGGCCAUGAAAAAUUCAUGGCCCGCCU ..--..............----................(((((((..(((..(((((((....)))..))))...)))....)))).))).(((((((((((.....))))))))))).. ( -31.30) >DroYak_CAF1 3855 120 + 1 CCAUCCAUCCCCACAACACAAGAACUCCAUCUAUUUUCUAUCCAAAUUGUCAGUUGGCCAGUUGGCCCCAACAUUGCAUGUCUUGGCAUACGUGGGCCAUGAAAAAUUCAUGGCCCGCCU ..........(((..(((..(((......)))....................(((((((....))))..)))......)))..))).....(((((((((((.....))))))))))).. ( -32.60) >consensus CC__CCUCCCCCACAACA____AACUCCAUCUAUUUUCUAUCCAAAUUGUCAGUUGGCCAGUUGGCUCCAACAUUGCAUGUCUUGGCAUACGUGGGCCAUGAAAAAUUCAUGGCCCGCCU ......................................(((((((..(((..(((((((....)))..))))...)))....)))).))).(((((((((((.....))))))))))).. (-31.30 = -31.30 + 0.00)

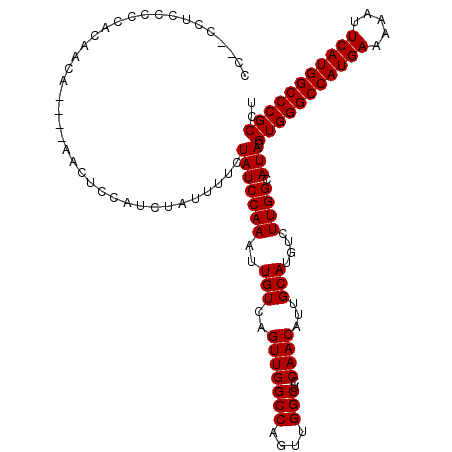
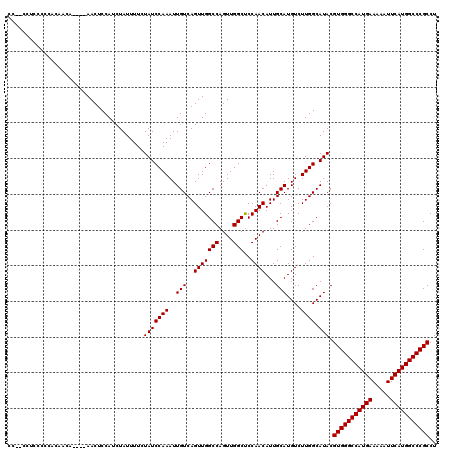
| Location | 14,762,106 – 14,762,219 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.09 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -32.43 |
| Energy contribution | -32.43 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14762106 113 - 20766785 AGGCGGGCCAUGAAUUUUUCAUGGCCCACGUAUGCCAAGACAUGCAAUGUUGGAGCCAACUGGCCAACUGACAAUUUGGAUAGAAAAUAGAUGGAGUU----UGGUCUG-CGGAGG--GG ....(((((((((.....))))))))).((((.((((((.(((....(((..(.(((....)))...)..)))((((.......))))..)))...))----)))).))-))....--.. ( -38.60) >DroSim_CAF1 2534 114 - 1 AGGCGGGCCAUGAAUUUUUCAUGGCCCACGUAUGCCAAGACAUGCAAUGUUGGAGCCAACUGGCCAACUGACAAUUUGGAUAGAAAAUAGAUGGAGUU----UGUUGAGGGGGAGG--GG .((((((((((((.....)))))))))..(((((......))))).........)))..((..((..((((((((..(..((.....))..)..)..)----)))).))..))..)--). ( -36.80) >DroYak_CAF1 3855 120 - 1 AGGCGGGCCAUGAAUUUUUCAUGGCCCACGUAUGCCAAGACAUGCAAUGUUGGGGCCAACUGGCCAACUGACAAUUUGGAUAGAAAAUAGAUGGAGUUCUUGUGUUGUGGGGAUGGAUGG ....(((((((((.....))))))))).(((.(.(((.(((((.((((((..(((((....))))..)..)))..)))..........(((......))).))))).))).))))..... ( -39.20) >consensus AGGCGGGCCAUGAAUUUUUCAUGGCCCACGUAUGCCAAGACAUGCAAUGUUGGAGCCAACUGGCCAACUGACAAUUUGGAUAGAAAAUAGAUGGAGUU____UGUUGUGGGGGAGG__GG .((((((((((((.....)))))))))..(((((......))))).........)))..(((.((((........)))).)))..................................... (-32.43 = -32.43 + -0.00)

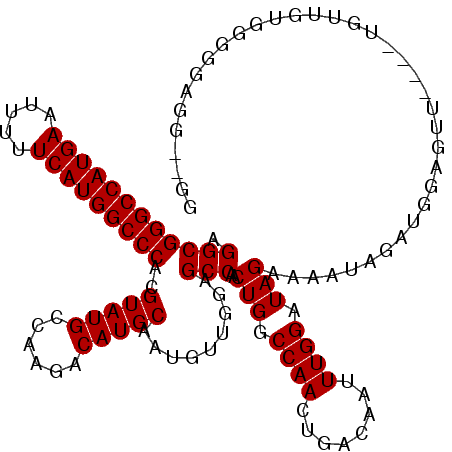
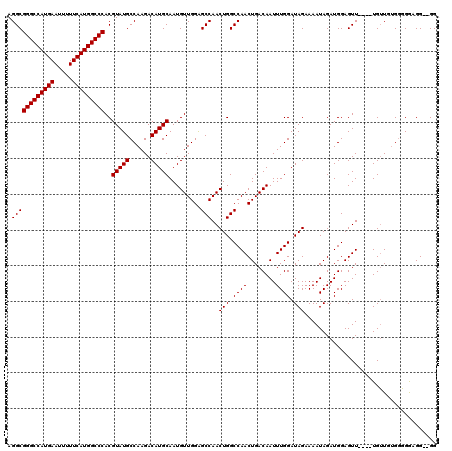
| Location | 14,762,139 – 14,762,259 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -45.32 |
| Consensus MFE | -45.14 |
| Energy contribution | -44.92 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.94 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14762139 120 + 20766785 UCCAAAUUGUCAGUUGGCCAGUUGGCUCCAACAUUGCAUGUCUUGGCAUACGUGGGCCAUGAAAAAUUCAUGGCCCGCCUUCAAUGUACUGCCACAUAAUUAGACUGUGGGGCUGCCAAA ............(..((((.((((....)))).(..((.(((((((((((((((((((((((.....))))))))))).......))).))))).......))))))..)))))..)... ( -44.52) >DroSim_CAF1 2568 120 + 1 UCCAAAUUGUCAGUUGGCCAGUUGGCUCCAACAUUGCAUGUCUUGGCAUACGUGGGCCAUGAAAAAUUCAUGGCCCGCCUUCAAUGUACUGCCACAUAAUUAGACUGUGGGGCUGCCAAA ............(..((((.((((....)))).(..((.(((((((((((((((((((((((.....))))))))))).......))).))))).......))))))..)))))..)... ( -44.52) >DroYak_CAF1 3895 120 + 1 UCCAAAUUGUCAGUUGGCCAGUUGGCCCCAACAUUGCAUGUCUUGGCAUACGUGGGCCAUGAAAAAUUCAUGGCCCGCCUUCAAUGUACUGCCACAUAAUUAGACUGUGGGGCUGCCAAA .............(((((.....((((((......(((.(((((((((((((((((((((((.....))))))))))).......))).))))).......)))))))))))))))))). ( -46.92) >consensus UCCAAAUUGUCAGUUGGCCAGUUGGCUCCAACAUUGCAUGUCUUGGCAUACGUGGGCCAUGAAAAAUUCAUGGCCCGCCUUCAAUGUACUGCCACAUAAUUAGACUGUGGGGCUGCCAAA .............(((((.....((((((......(((.(((((((((((((((((((((((.....))))))))))).......))).))))).......)))))))))))))))))). (-45.14 = -44.92 + -0.22)

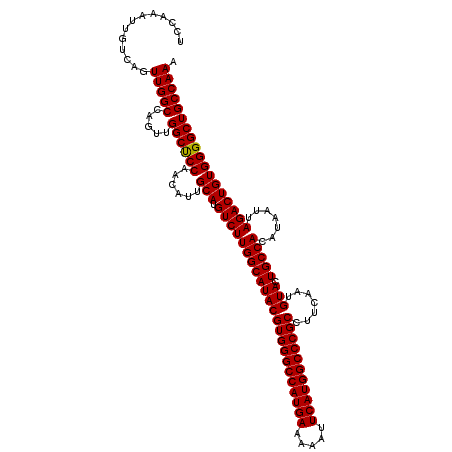
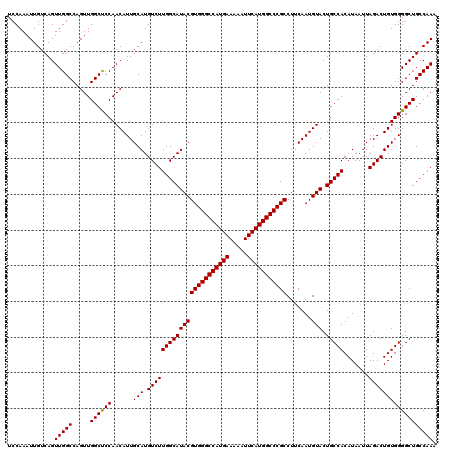
| Location | 14,762,139 – 14,762,259 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -44.97 |
| Consensus MFE | -44.27 |
| Energy contribution | -44.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14762139 120 - 20766785 UUUGGCAGCCCCACAGUCUAAUUAUGUGGCAGUACAUUGAAGGCGGGCCAUGAAUUUUUCAUGGCCCACGUAUGCCAAGACAUGCAAUGUUGGAGCCAACUGGCCAACUGACAAUUUGGA (((((((((((((((.........)))))(((....)))..)))(((((((((.....))))))))).....))))))).....((((((..(.(((....)))...)..)))..))).. ( -43.90) >DroSim_CAF1 2568 120 - 1 UUUGGCAGCCCCACAGUCUAAUUAUGUGGCAGUACAUUGAAGGCGGGCCAUGAAUUUUUCAUGGCCCACGUAUGCCAAGACAUGCAAUGUUGGAGCCAACUGGCCAACUGACAAUUUGGA (((((((((((((((.........)))))(((....)))..)))(((((((((.....))))))))).....))))))).....((((((..(.(((....)))...)..)))..))).. ( -43.90) >DroYak_CAF1 3895 120 - 1 UUUGGCAGCCCCACAGUCUAAUUAUGUGGCAGUACAUUGAAGGCGGGCCAUGAAUUUUUCAUGGCCCACGUAUGCCAAGACAUGCAAUGUUGGGGCCAACUGGCCAACUGACAAUUUGGA (((((((((((((((.........)))))(((....)))..)))(((((((((.....))))))))).....))))))).....((((((..(((((....))))..)..)))..))).. ( -47.10) >consensus UUUGGCAGCCCCACAGUCUAAUUAUGUGGCAGUACAUUGAAGGCGGGCCAUGAAUUUUUCAUGGCCCACGUAUGCCAAGACAUGCAAUGUUGGAGCCAACUGGCCAACUGACAAUUUGGA (((((((((((((((.........)))))(((....)))..)))(((((((((.....))))))))).....))))))).....((((((..(.(((....)))...)..)))..))).. (-44.27 = -44.27 + 0.00)

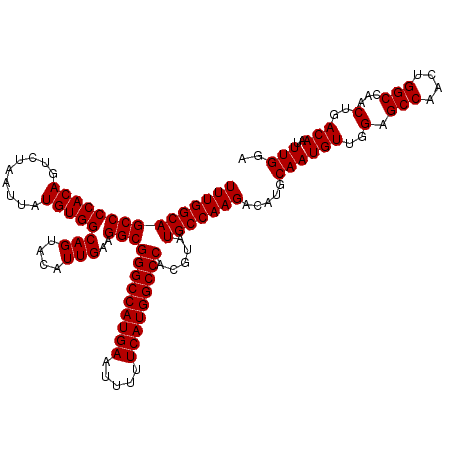
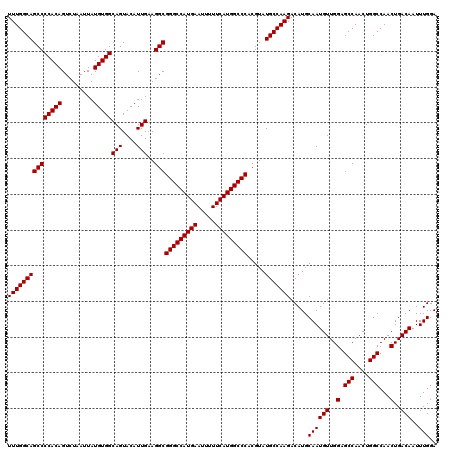
| Location | 14,762,179 – 14,762,299 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.38 |
| Mean single sequence MFE | -42.44 |
| Consensus MFE | -39.04 |
| Energy contribution | -39.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.10 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14762179 120 + 20766785 UCUUGGCAUACGUGGGCCAUGAAAAAUUCAUGGCCCGCCUUCAAUGUACUGCCACAUAAUUAGACUGUGGGGCUGCCAAAAAAAUAAAGGUUGUCCUGAAAGCAGGAUUGUUAAUAGAUA ..((((((...(((((((((((.....)))))))))))............(((.((((.......)))).))))))))).............((((((....))))))............ ( -41.90) >DroSim_CAF1 2608 119 + 1 UCUUGGCAUACGUGGGCCAUGAAAAAUUCAUGGCCCGCCUUCAAUGUACUGCCACAUAAUUAGACUGUGGGGCUGCCAAA-AAAUAAAGGUUGUCCUGAAAGCGGGAUUGUUAAUAGAUA ..((((((...(((((((((((.....)))))))))))............(((.((((.......)))).))))))))).-...........((((((....))))))............ ( -41.20) >DroYak_CAF1 3935 119 + 1 UCUUGGCAUACGUGGGCCAUGAAAAAUUCAUGGCCCGCCUUCAAUGUACUGCCACAUAAUUAGACUGUGGGGCUGCCAAA-AAAUGGAGGUUGUCCUCAAAGCCUGAUUGUUAAUAGAUA ...(((((((((((((((((((.....))))))))))).......))).))))).((((((((.((.((((((.(((...-.......))).)))).)).)).))))))))......... ( -44.21) >consensus UCUUGGCAUACGUGGGCCAUGAAAAAUUCAUGGCCCGCCUUCAAUGUACUGCCACAUAAUUAGACUGUGGGGCUGCCAAA_AAAUAAAGGUUGUCCUGAAAGCAGGAUUGUUAAUAGAUA ...(((((((((((((((((((.....))))))))))).......))).))))).((((((...((..(((((.(((...........))).)))))...))...))))))......... (-39.04 = -39.04 + -0.00)


| Location | 14,762,179 – 14,762,299 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.38 |
| Mean single sequence MFE | -39.43 |
| Consensus MFE | -36.10 |
| Energy contribution | -36.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.03 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14762179 120 - 20766785 UAUCUAUUAACAAUCCUGCUUUCAGGACAACCUUUAUUUUUUUGGCAGCCCCACAGUCUAAUUAUGUGGCAGUACAUUGAAGGCGGGCCAUGAAUUUUUCAUGGCCCACGUAUGCCAAGA .............(((((....)))))............((((((((((((((((.........)))))(((....)))..)))(((((((((.....))))))))).....)))))))) ( -41.00) >DroSim_CAF1 2608 119 - 1 UAUCUAUUAACAAUCCCGCUUUCAGGACAACCUUUAUUU-UUUGGCAGCCCCACAGUCUAAUUAUGUGGCAGUACAUUGAAGGCGGGCCAUGAAUUUUUCAUGGCCCACGUAUGCCAAGA .............(((........)))...........(-(((((((((((((((.........)))))(((....)))..)))(((((((((.....))))))))).....)))))))) ( -37.30) >DroYak_CAF1 3935 119 - 1 UAUCUAUUAACAAUCAGGCUUUGAGGACAACCUCCAUUU-UUUGGCAGCCCCACAGUCUAAUUAUGUGGCAGUACAUUGAAGGCGGGCCAUGAAUUUUUCAUGGCCCACGUAUGCCAAGA ......................((((....))))....(-(((((((((((((((.........)))))(((....)))..)))(((((((((.....))))))))).....)))))))) ( -40.00) >consensus UAUCUAUUAACAAUCCCGCUUUCAGGACAACCUUUAUUU_UUUGGCAGCCCCACAGUCUAAUUAUGUGGCAGUACAUUGAAGGCGGGCCAUGAAUUUUUCAUGGCCCACGUAUGCCAAGA .......................(((....))).......(((((((((((((((.........)))))(((....)))..)))(((((((((.....))))))))).....))))))). (-36.10 = -36.10 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:08 2006