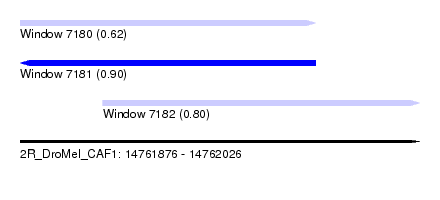
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,761,876 – 14,762,026 |
| Length | 150 |
| Max. P | 0.900102 |

| Location | 14,761,876 – 14,761,987 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.00 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -23.32 |
| Energy contribution | -23.43 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14761876 111 + 20766785 AGUUAUUAUUCCCAUG-----UUUUCGUU-UUGU---GGGGUGGAACAUAUGGCCUUUUUUUCAGGUGGGUCAUUCCAUGGCCCAUUUAUGGAAUUAUGCCAUGGCAUAAAAAUUUGAUG .(((.....(((((((-----........-.)))---))))...))).((((((..(..(((((.(((((((((...)))))))))...)))))..).))))))................ ( -30.60) >DroSim_CAF1 2304 110 + 1 AGUUAUUUUU-CCAUG-----UUUUCGUUUUUGG---GGGGUGGAACAUAUGGCCUUU-UUUCAGGUGGGUCAUUCCAUGGCCCAUUUAUGGAAUUAUGCCAUGGCAUAAAAAUUUGAUG .(((((..((-((((.-----((..((....)).---.))))))))...)))))....-(((((.(((((((((...)))))))))...)))))((((((....)))))).......... ( -31.70) >DroYak_CAF1 3619 117 + 1 AGUUAUUUUUCCGGUUUUGGUUUUUGGUU-UUUUGGGGGGUUGGAACAUAUGGCCCU--UUUCAGGUGGGUCAUUCCAUGGCCCAUUUAUGGAAUUAUGCCAUGGCAUAAAAAUUUGAUG .((((..(((...((..((((...(((((-(..((((((((((((....(((((((.--........)))))))))))..)))).))))..)))))).))))..))....)))..)))). ( -32.70) >consensus AGUUAUUUUUCCCAUG_____UUUUCGUU_UUGU___GGGGUGGAACAUAUGGCCUUU_UUUCAGGUGGGUCAUUCCAUGGCCCAUUUAUGGAAUUAUGCCAUGGCAUAAAAAUUUGAUG .....................................((((((((....(((((((...........))))))))))))..)))..........((((((....)))))).......... (-23.32 = -23.43 + 0.11)

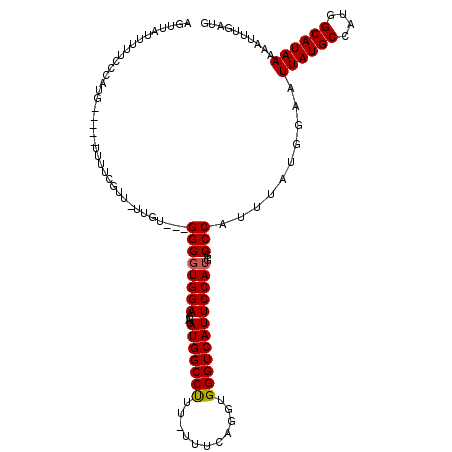
| Location | 14,761,876 – 14,761,987 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.00 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -17.49 |
| Energy contribution | -17.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14761876 111 - 20766785 CAUCAAAUUUUUAUGCCAUGGCAUAAUUCCAUAAAUGGGCCAUGGAAUGACCCACCUGAAAAAAAGGCCAUAUGUUCCACCCC---ACAA-AACGAAAA-----CAUGGGAAUAAUAACU ..........((((((....))))))((((((...((((...((((((......(((.......)))......)))))).)))---)(..-...)....-----.))))))......... ( -22.90) >DroSim_CAF1 2304 110 - 1 CAUCAAAUUUUUAUGCCAUGGCAUAAUUCCAUAAAUGGGCCAUGGAAUGACCCACCUGAAA-AAAGGCCAUAUGUUCCACCCC---CCAAAAACGAAAA-----CAUGG-AAAAAUAACU ..........((((((....))))))((((((...((((...((((((......(((....-..)))......))))))...)---))).....(....-----)))))-))........ ( -24.90) >DroYak_CAF1 3619 117 - 1 CAUCAAAUUUUUAUGCCAUGGCAUAAUUCCAUAAAUGGGCCAUGGAAUGACCCACCUGAAA--AGGGCCAUAUGUUCCAACCCCCCAAAA-AACCAAAAACCAAAACCGGAAAAAUAACU ..........((((((....)))))).........((((...((((((......(((....--))).......))))))....))))...-.........((......)).......... ( -23.22) >consensus CAUCAAAUUUUUAUGCCAUGGCAUAAUUCCAUAAAUGGGCCAUGGAAUGACCCACCUGAAA_AAAGGCCAUAUGUUCCACCCC___ACAA_AACGAAAA_____CAUGGGAAAAAUAACU ..........((((((....))))))..........(((...((((((......(((.......)))......)))))).)))..................................... (-17.49 = -17.27 + -0.22)

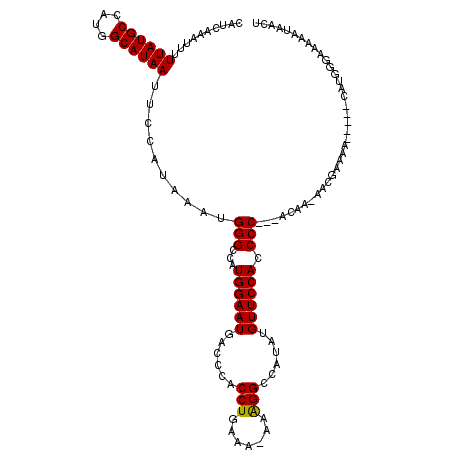
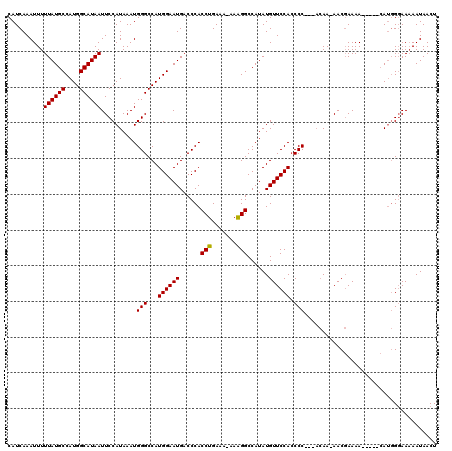
| Location | 14,761,907 – 14,762,026 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.09 |
| Mean single sequence MFE | -33.63 |
| Consensus MFE | -30.77 |
| Energy contribution | -30.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14761907 119 + 20766785 GUGGAACAUAUGGCCUUUUUUUCAGGUGGGUCAUUCCAUGGCCCAUUUAUGGAAUUAUGCCAUGGCAUAAAAAUUUGAUGAGCUUCAACUUCACAAGUGCCUGGAUCCCA-AUGAAAAGU .................(((((((..((((....((((.((((((....)))..((((((....))))))..(((((.((((.......)))))))))))))))).))))-.))))))). ( -32.40) >DroSim_CAF1 2335 119 + 1 GUGGAACAUAUGGCCUUU-UUUCAGGUGGGUCAUUCCAUGGCCCAUUUAUGGAAUUAUGCCAUGGCAUAAAAAUUUGAUGAGCUUCAACUUCACAAGUGCCUGGAUCCCAAAUGAAAAGU ................((-(((((..((((....((((.((((((....)))..((((((....))))))..(((((.((((.......)))))))))))))))).))))..))))))). ( -34.00) >DroYak_CAF1 3658 117 + 1 UUGGAACAUAUGGCCCU--UUUCAGGUGGGUCAUUCCAUGGCCCAUUUAUGGAAUUAUGCCAUGGCAUAAAAAUUUGAUGAGCUUCCACUUCACAAGUGCCAGGAUCCCA-AUGAAAAGU ...............((--(((((..((((....(((.(((((((....)))..((((((....))))))..(((((.((((.......)))))))))))))))).))))-.))))))). ( -34.50) >consensus GUGGAACAUAUGGCCUUU_UUUCAGGUGGGUCAUUCCAUGGCCCAUUUAUGGAAUUAUGCCAUGGCAUAAAAAUUUGAUGAGCUUCAACUUCACAAGUGCCUGGAUCCCA_AUGAAAAGU ..(((.(..(((((.....(((((.(((((((((...)))))))))...)))))....)))))(((((......(((.((((.......))))))))))))..).)))............ (-30.77 = -30.77 + 0.00)

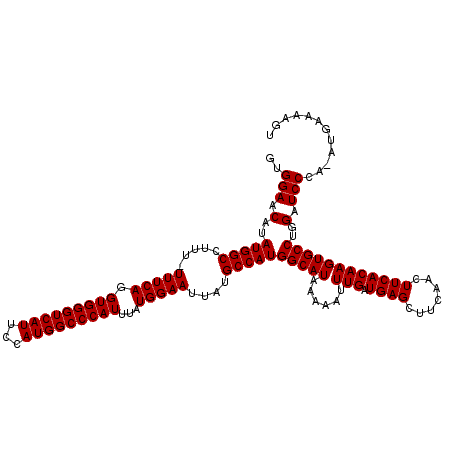
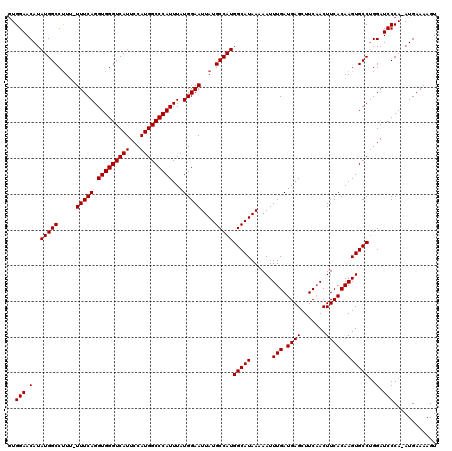
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:03 2006