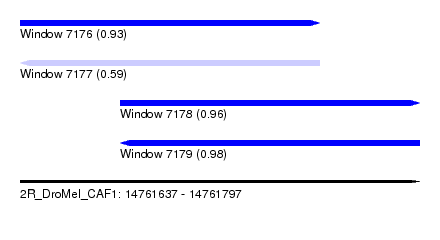
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,761,637 – 14,761,797 |
| Length | 160 |
| Max. P | 0.982499 |

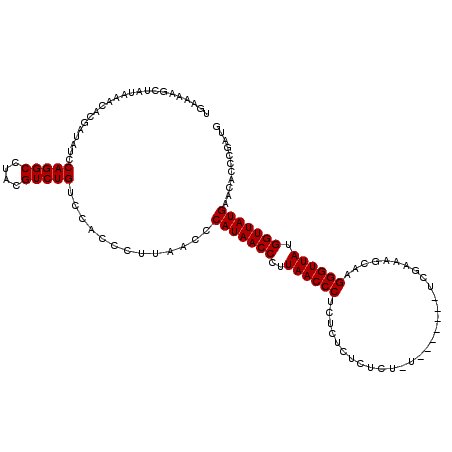
| Location | 14,761,637 – 14,761,757 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.57 |
| Mean single sequence MFE | -23.61 |
| Consensus MFE | -19.71 |
| Energy contribution | -19.71 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14761637 120 + 20766785 UGAAAAGCUAUAAACACGAUAACCAGGCCUACGUCUGUCCACCCUUAACCCAUAACCCUUAACCCUCUCUCUCUCUCUCUCUCAUUCGAAAGCAAGGGUUAUGGUUAUGAACACCCGAUG ................((.....(((((....))))).............(((((((..(((((((..((.((..............)).))..))))))).)))))))......))... ( -24.34) >DroSim_CAF1 2070 114 + 1 UGAAAAGCUAUAAACACGAUAUCCAGGCCUACGUCUGUCCACCCUUAACCCAUAACCCUUAACCCUCUCUCUCUCUUUA------UCGAAAGCAAGGGUUAUGGUUAUGAACACCCGAUG ................((.....(((((....))))).............(((((((..(((((((..((.((......------..)).))..))))))).)))))))......))... ( -25.60) >DroYak_CAF1 3398 102 + 1 UGAAAAGCUAUAAACACGAUAUCCAGGCCUACGUCUGUCCACCCUUAACCCAUAACCCUUAACCCUCUCUCUCUC------------------AUGGGUUAUGGUUAUGAACACCCAAUG .......................(((((....))))).............(((((((..((((((..........------------------..)))))).)))))))........... ( -20.90) >consensus UGAAAAGCUAUAAACACGAUAUCCAGGCCUACGUCUGUCCACCCUUAACCCAUAACCCUUAACCCUCUCUCUCUCU_U_______UCGAAAGCAAGGGUUAUGGUUAUGAACACCCGAUG .......................(((((....))))).............(((((((..((((((..............................)))))).)))))))........... (-19.71 = -19.71 + 0.00)

| Location | 14,761,637 – 14,761,757 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.57 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -28.09 |
| Energy contribution | -29.87 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14761637 120 - 20766785 CAUCGGGUGUUCAUAACCAUAACCCUUGCUUUCGAAUGAGAGAGAGAGAGAGAGAGGGUUAAGGGUUAUGGGUUAAGGGUGGACAGACGUAGGCCUGGUUAUCGUGUUUAUAGCUUUUCA ((((....(..(((((((.((((((((.(((((...(....)...)))))...))))))))..)))))))..)....))))((.((..((((((.((.....)).))))))..))..)). ( -35.30) >DroSim_CAF1 2070 114 - 1 CAUCGGGUGUUCAUAACCAUAACCCUUGCUUUCGA------UAAAGAGAGAGAGAGGGUUAAGGGUUAUGGGUUAAGGGUGGACAGACGUAGGCCUGGAUAUCGUGUUUAUAGCUUUUCA ((((....(..(((((((.((((((((.(((((..------....)))))...))))))))..)))))))..)....))))((.((..((((((.((.....)).))))))..))..)). ( -36.00) >DroYak_CAF1 3398 102 - 1 CAUUGGGUGUUCAUAACCAUAACCCAU------------------GAGAGAGAGAGGGUUAAGGGUUAUGGGUUAAGGGUGGACAGACGUAGGCCUGGAUAUCGUGUUUAUAGCUUUUCA .....(((((((....(((((((((.(------------------((...........))).))))))))).....((((..((....))..)))))))))))................. ( -26.40) >consensus CAUCGGGUGUUCAUAACCAUAACCCUUGCUUUCGA_______A_AGAGAGAGAGAGGGUUAAGGGUUAUGGGUUAAGGGUGGACAGACGUAGGCCUGGAUAUCGUGUUUAUAGCUUUUCA ((((....(..(((((((.((((((((.(((((..............))))).))))))))..)))))))..)....))))((.((..((((((.((.....)).))))))..))..)). (-28.09 = -29.87 + 1.78)

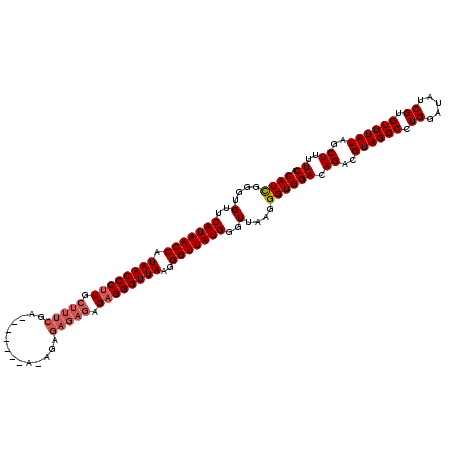
| Location | 14,761,677 – 14,761,797 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.14 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -25.81 |
| Energy contribution | -25.81 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.82 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14761677 120 + 20766785 ACCCUUAACCCAUAACCCUUAACCCUCUCUCUCUCUCUCUCUCAUUCGAAAGCAAGGGUUAUGGUUAUGAACACCCGAUGUGUGGGUUAACCAAAAGGCAUUUUCAAUAAAUACCAAUUA ....(((((((((((((..(((((((..((.((..............)).))..))))))).))))))..((((.....)))))))))))......((.((((.....)))).))..... ( -29.64) >DroSim_CAF1 2110 114 + 1 ACCCUUAACCCAUAACCCUUAACCCUCUCUCUCUCUUUA------UCGAAAGCAAGGGUUAUGGUUAUGAACACCCGAUGUGUGGGUUAACCAAAAGGCAUUUUCAAUAAAUACCAAUUA ....(((((((((((((..(((((((..((.((......------..)).))..))))))).))))))..((((.....)))))))))))......((.((((.....)))).))..... ( -30.90) >DroYak_CAF1 3438 102 + 1 ACCCUUAACCCAUAACCCUUAACCCUCUCUCUCUC------------------AUGGGUUAUGGUUAUGAACACCCAAUGUGUGGGUUAACCAAAAGGCAUUUUCAAUAAAUACCAAUUA ....(((((((((((((..((((((..........------------------..)))))).))))))..((((.....)))))))))))......((.((((.....)))).))..... ( -27.00) >consensus ACCCUUAACCCAUAACCCUUAACCCUCUCUCUCUCU_U_______UCGAAAGCAAGGGUUAUGGUUAUGAACACCCGAUGUGUGGGUUAACCAAAAGGCAUUUUCAAUAAAUACCAAUUA ....(((((((((((((..((((((..............................)))))).))))))..((((.....)))))))))))......((.((((.....)))).))..... (-25.81 = -25.81 + 0.00)



| Location | 14,761,677 – 14,761,797 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.14 |
| Mean single sequence MFE | -37.93 |
| Consensus MFE | -34.14 |
| Energy contribution | -36.14 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.15 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14761677 120 - 20766785 UAAUUGGUAUUUAUUGAAAAUGCCUUUUGGUUAACCCACACAUCGGGUGUUCAUAACCAUAACCCUUGCUUUCGAAUGAGAGAGAGAGAGAGAGAGGGUUAAGGGUUAUGGGUUAAGGGU (((..(((((((.....)))))))..))).(((((((((((.....))))..((((((.((((((((.(((((...(....)...)))))...))))))))..))))))))))))).... ( -41.00) >DroSim_CAF1 2110 114 - 1 UAAUUGGUAUUUAUUGAAAAUGCCUUUUGGUUAACCCACACAUCGGGUGUUCAUAACCAUAACCCUUGCUUUCGA------UAAAGAGAGAGAGAGGGUUAAGGGUUAUGGGUUAAGGGU (((..(((((((.....)))))))..))).(((((((((((.....))))..((((((.((((((((.(((((..------....)))))...))))))))..))))))))))))).... ( -41.70) >DroYak_CAF1 3438 102 - 1 UAAUUGGUAUUUAUUGAAAAUGCCUUUUGGUUAACCCACACAUUGGGUGUUCAUAACCAUAACCCAU------------------GAGAGAGAGAGGGUUAAGGGUUAUGGGUUAAGGGU (((..(((((((.....)))))))..))).(((((((((((.....))))..((((((.((((((..------------------..........))))))..))))))))))))).... ( -31.10) >consensus UAAUUGGUAUUUAUUGAAAAUGCCUUUUGGUUAACCCACACAUCGGGUGUUCAUAACCAUAACCCUUGCUUUCGA_______A_AGAGAGAGAGAGGGUUAAGGGUUAUGGGUUAAGGGU (((..(((((((.....)))))))..))).(((((((((((.....))))..((((((.((((((((.(((((..............))))).))))))))..))))))))))))).... (-34.14 = -36.14 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:00 2006