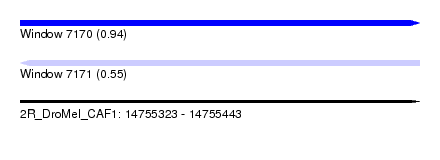
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,755,323 – 14,755,443 |
| Length | 120 |
| Max. P | 0.938810 |

| Location | 14,755,323 – 14,755,443 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -22.75 |
| Energy contribution | -21.53 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.938810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
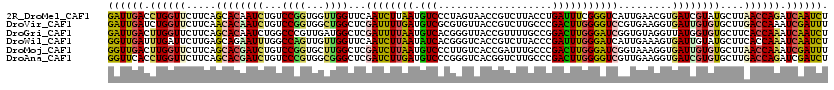
>2R_DroMel_CAF1 14755323 120 + 20766785 AGAUUGAUCUGGUUAAGCAUACGAUCACGUUCAAUGACCCGAAAUCAGGUAAGACGGUUACUAGGGACAUUAAGAUUGAACCAACCACCGGACAGAUUGUGCUGAAGAACCAGGUCAAUC .(((((((((((((...(((((((((..((((....(((.(....).))).....((((....((..((.......))..))))))...)))).))))))).))...))))))))))))) ( -39.00) >DroVir_CAF1 54557 120 + 1 AAAUCGAUUUGGUCAAGCACACAAUCACCUUCACGGACCCCAAGUCGGGCAAGACGGUAACACGCGACAUCAAAAUCGAGCCAGCCACCGGACAGAUUGUGUUGAAGAACCAGAUCAAUC .....((((((((....(((((((((....((.(((.......(((......)))(((......(((........))).....))).)))))..))))))).))....)))))))).... ( -33.00) >DroGri_CAF1 63393 120 + 1 AGAUUGAUUUGGUGAAGCACACCAUAACCUACACCGAUCCCAAGUCCGGCAAAACGGUAACCCGUGACAUUAAAAUCGAGCCAUCAACGGGCCAGAUUGUGCUGAAGAACCAAGUCAAUC .((((((((((((..(((((.......................((((((............))).))).....((((..(((.......)))..))))))))).....)))))))))))) ( -36.20) >DroWil_CAF1 79523 120 + 1 AGAUUGAUUUGGUGAAGCAUACAAUCACUUUCAAUGAUCCCAAAUCGGGUAAGACGGUGACCCGUGAUAUUAAGAUUGAACCAACAACUGGCCAAAUUCUGCUCAAGAAUCAAAUCAACC ...((((((((((..((((..(((((............(((.....)))....((((....))))........)))))..(((.....)))........)))).....)))))))))).. ( -26.50) >DroMoj_CAF1 67792 120 + 1 AAAUCGAUUUGGUUAAGCACACAAUCACCUUUACCGAUCCCAAGUCGGGCAAAUCGGUGACAAGGGACAUUAAGAUCGAGCCAAGCACCGGACAGAUCGUGCUGAAGAACCAAGUCAACC .....(((((((((.(((((....((.((((((((((((((.....)))...))))))))..)))))......((((...((.......))...)))))))))....))))))))).... ( -43.70) >DroAna_CAF1 52051 120 + 1 AGAUCGAUCUGGUCAAGCACACGAUCACCUUCAACGACCCCAAGUCGGGCAAGACCGUGACCCGGGACAUCAAGAUCGAGCCCGCCACGGGACAGAUCGUGCUGAAGAACCAGGUGAACC ...((.(((((((....(((((((((..(((...((((.....))))...))).(((((...((((...((......)).)))).)))))....))))))).))....)))))))))... ( -43.00) >consensus AGAUCGAUUUGGUCAAGCACACAAUCACCUUCAACGACCCCAAGUCGGGCAAGACGGUGACCCGGGACAUUAAGAUCGAGCCAACCACCGGACAGAUUGUGCUGAAGAACCAAGUCAACC .....((((((((..(((((..................(((.....)))........................((((...((.......))...))))))))).....)))))))).... (-22.75 = -21.53 + -1.22)

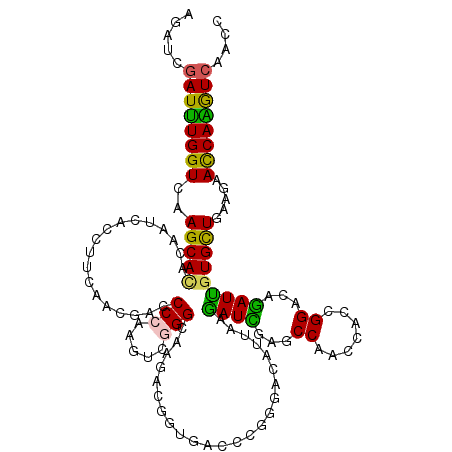
| Location | 14,755,323 – 14,755,443 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -38.26 |
| Consensus MFE | -23.71 |
| Energy contribution | -23.24 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.49 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14755323 120 - 20766785 GAUUGACCUGGUUCUUCAGCACAAUCUGUCCGGUGGUUGGUUCAAUCUUAAUGUCCCUAGUAACCGUCUUACCUGAUUUCGGGUCAUUGAACGUGAUCGUAUGCUUAACCAGAUCAAUCU ((((((.((((((....(((((((((........)))))(.(((...((((((.(((..((((.....))))..(....)))).))))))...))).)...)))).)))))).)))))). ( -34.00) >DroVir_CAF1 54557 120 - 1 GAUUGAUCUGGUUCUUCAACACAAUCUGUCCGGUGGCUGGCUCGAUUUUGAUGUCGCGUGUUACCGUCUUGCCCGACUUGGGGUCCGUGAAGGUGAUUGUGUGCUUGACCAAAUCGAUUU (((((((.(((((.....((((((((...((((((((..((.((((......)))).)))))))).((..((((......))))....)).)).))))))))....))))).))))))). ( -42.00) >DroGri_CAF1 63393 120 - 1 GAUUGACUUGGUUCUUCAGCACAAUCUGGCCCGUUGAUGGCUCGAUUUUAAUGUCACGGGUUACCGUUUUGCCGGACUUGGGAUCGGUGUAGGUUAUGGUGUGCUUCACCAAAUCAAUCU ((((((.(((((.....((((((..((((((((....((((..(((......)))((((....))))...))))....)))).))))............))))))..))))).)))))). ( -39.24) >DroWil_CAF1 79523 120 - 1 GGUUGAUUUGAUUCUUGAGCAGAAUUUGGCCAGUUGUUGGUUCAAUCUUAAUAUCACGGGUCACCGUCUUACCCGAUUUGGGAUCAUUGAAAGUGAUUGUAUGCUUCACCAAAUCAAUCU ((((((((((......(((((......((((((...))))))(((((........((((....))))....(((.....)))............)))))..)))))...)))))))))). ( -29.50) >DroMoj_CAF1 67792 120 - 1 GGUUGACUUGGUUCUUCAGCACGAUCUGUCCGGUGCUUGGCUCGAUCUUAAUGUCCCUUGUCACCGAUUUGCCCGACUUGGGAUCGGUAAAGGUGAUUGUGUGCUUAACCAAAUCGAUUU ((((((.((((((....(((((((((.(((........)))..)))).....(((((((...((((((...(((.....))))))))).)))).)))...))))).)))))).)))))). ( -37.60) >DroAna_CAF1 52051 120 - 1 GGUUCACCUGGUUCUUCAGCACGAUCUGUCCCGUGGCGGGCUCGAUCUUGAUGUCCCGGGUCACGGUCUUGCCCGACUUGGGGUCGUUGAAGGUGAUCGUGUGCUUGACCAGAUCGAUCU ((((...((((((.....((((((((.(.((((...)))))...((((((((((((((((((..((.....)).))))))))).)))).)))))))))))))....))))))...)))). ( -47.20) >consensus GAUUGACCUGGUUCUUCAGCACAAUCUGUCCGGUGGUUGGCUCGAUCUUAAUGUCCCGGGUCACCGUCUUGCCCGACUUGGGAUCGGUGAAGGUGAUUGUGUGCUUAACCAAAUCAAUCU ((((((.((((((.....((((((((...((((...))))...((((((((.(((...................))))))))))).........))))))))....)))))).)))))). (-23.71 = -23.24 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:53 2006