
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,703,426 – 14,703,568 |
| Length | 142 |
| Max. P | 0.764717 |

| Location | 14,703,426 – 14,703,543 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.46 |
| Mean single sequence MFE | -24.16 |
| Consensus MFE | -12.00 |
| Energy contribution | -13.88 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
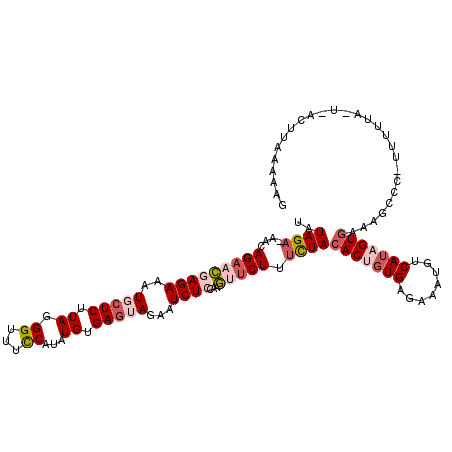
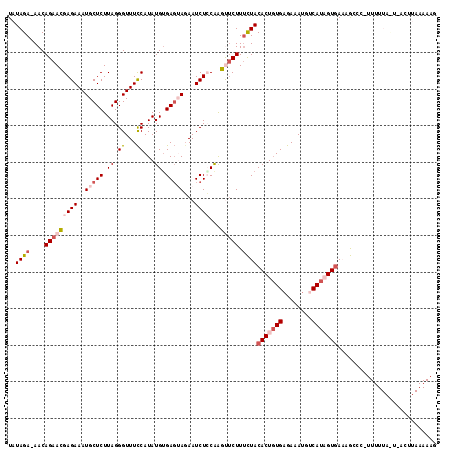
>2R_DroMel_CAF1 14703426 117 + 20766785 UAUAGAAAACAGAACGAGAAAUGCUCUUAGGGUUUCCAUAUGUGAGUACAAUCUGCAAGUUCUUUCUACACUCUGAGAAAUGUCAUAGUGAUUGCCCAUUUUUACUCACUUAAAAAG ...............(.(((((.(.....).))))))....(((((((.((...(((((......)).((((.(((......))).))))..)))....)).)))))))........ ( -19.90) >DroSec_CAF1 8852 115 + 1 UAUAGA-AACAGAACGAGAAAUGCUCUUAGGGUUUCCAUAUGUGAGUAGAAUCUCCAAGUUCUUUCUACACUGUGAGAAAUGUCAUAGUGAUAGCCC-UUUUUACUAACUUAAAAAC ..((((-(..(((((((((..(((((.((((....)).))...)))))...))))...))))))))))((((((((......)))))))).......-................... ( -26.10) >DroSim_CAF1 11286 115 + 1 UAUAGA-AACAGAACGAGAAAUGCUCUUAGGGUUUCCAUAUGUGAGUAGAAUCUCCGAGUUCUUUCUACACUGUGAGAAACGUCAUAGUGAAAGCCC-UUUUUAUUCACUUAAAAAG ..((((-(..(((((((((..(((((.((((....)).))...)))))...))))...))))))))))((((((((......))))))))......(-((((((......))))))) ( -29.10) >DroEre_CAF1 8731 92 + 1 UAUAGA-AACAGAAAGAGAAAUGCUCUUAGGGUUUCCAUAUGGGAGGAGAAUCUCCAGUCGCUUUUUACACUGUGAGAAAUG-CAUUGUGAAAA----------------------- (((.((-(((...(((((.....)))))...))))).)))..((((......)))).......(((((((.(((.......)-)).))))))).----------------------- ( -21.30) >DroYak_CAF1 9263 93 + 1 UAUAGA-AACAGAACGAGAAAUGCUCUUAGGGUUUUCAUAUGUGACGAGAAUCUACAAGCUCUUGUUACACUGUGAGAAAUC-CAUUGUAAAAA----------------------A ......-.((((...(((.....)))...(((((((((((((((((((((..........)))))))))).)))))).))))-).)))).....----------------------. ( -24.40) >consensus UAUAGA_AACAGAACGAGAAAUGCUCUUAGGGUUUCCAUAUGUGAGUAGAAUCUCCAAGUUCUUUCUACACUGUGAGAAAUGUCAUAGUGAAAGCCC_UUUUUA_U_ACUUAAAAAG ..((((....(((((((((..(((((.((.((...))...)).)))))...))))...))))).))))(((((((........)))))))........................... (-12.00 = -13.88 + 1.88)

| Location | 14,703,463 – 14,703,568 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -17.80 |
| Consensus MFE | -15.95 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
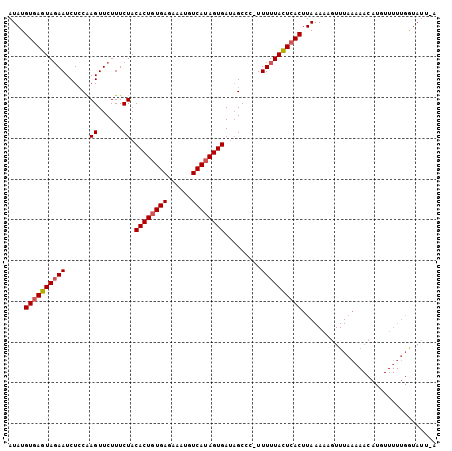
>2R_DroMel_CAF1 14703463 105 + 20766785 AUAUGUGAGUACAAUCUGCAAGUUCUUUCUACACUCUGAGAAAUGUCAUAGUGAUUGCCCAUUUUUACUCACUUAAAAAGUUUAAAAACAUGUUUUUGGUAUUAA ....(((((((.((...(((((......)).((((.(((......))).))))..)))....)).))))))).........(((((((....)))))))...... ( -17.00) >DroSec_CAF1 8888 88 + 1 AUAUGUGAGUAGAAUCUCCAAGUUCUUUCUACACUGUGAGAAAUGUCAUAGUGAUAGCCC-UUUUUACUAACUUAAAAACUUUCUG------------AUA---- .....((((((((((......)))))..(((((((((((......)))))))).)))...-.........)))))...........------------...---- ( -14.80) >DroSim_CAF1 11322 104 + 1 AUAUGUGAGUAGAAUCUCCGAGUUCUUUCUACACUGUGAGAAACGUCAUAGUGAAAGCCC-UUUUUAUUCACUUAAAAAGUUUAAAAACAUGUUUUUGGUAUUUA ....(((((((((......(((..(((((...(((((((......))))))))))))..)-))))))))))).....(((((((((((....))))))).)))). ( -21.60) >consensus AUAUGUGAGUAGAAUCUCCAAGUUCUUUCUACACUGUGAGAAAUGUCAUAGUGAUAGCCC_UUUUUACUCACUUAAAAAGUUUAAAAACAUGUUUUUGGUAUU_A ....((((((((((......((......)).((((((((......)))))))).........))))))))))................................. (-15.95 = -16.73 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:40 2006