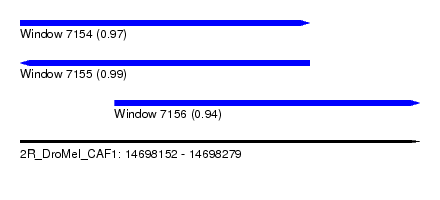
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,698,152 – 14,698,279 |
| Length | 127 |
| Max. P | 0.985585 |

| Location | 14,698,152 – 14,698,244 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 95.34 |
| Mean single sequence MFE | -23.78 |
| Consensus MFE | -20.90 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14698152 92 + 20766785 GACAGUAUGUGUUUAUGUUUGCUGAUUUGCUGCUUGUUUGGUUUCGGUUUUCACACAAAUUGCAAUU------GCAAUUGCAGUUGGCUCUGCUGCUA ..((((((((((....(...(((((...(((........))).)))))...)))))).....(((((------((....)))))))....)))))... ( -21.60) >DroSec_CAF1 3664 92 + 1 GACAGUAUGUGUUUAUGUUUGCUGAUUUGCUGCUUGUUUGGUUUCGGUUCUCACACAAAUUGCAAUU------GCAAUUGCAGUUGGCUCGGCUGCUA .......(((((....(...(((((...(((........))).))))).)..)))))((((((....------))))))(((((((...))))))).. ( -22.20) >DroSim_CAF1 6008 92 + 1 GACAGUAUGUGUUUAUGUUUGCUGAUUUGCUGCUUGUUUGGUUUCGGUUUUCACACAAAUUGCAAUU------GCAAUUGCAGUUGGCUCUGCUGCUA ..((((((((((....(...(((((...(((........))).)))))...)))))).....(((((------((....)))))))....)))))... ( -21.60) >DroEre_CAF1 3504 92 + 1 GACAGUGUGUGUUUAUGUUUGCUGAUUUGCUGCUUGUUUGGUUUCGGUUUUCACACAAAUUGCAAUU------GCAAUUGCAGUUUGCUCUGCUGCUA .....((((((.........(((((...(((........))).)))))...))))))((((((....------))))))(((((.......))))).. ( -22.90) >DroYak_CAF1 3621 98 + 1 GACAGUGUGUGUUUAUGUUUGCUGAUUUGCUGCUUGUUUGGUUUCGGUUUUCACACAAAUUGCAAUUGCAAUUGCAAUUGCAGUUUGCUCUGCUGCUA ..(((((((((.........(((((...(((........))).)))))...)))))(((((((((((((....))))))))))))).....))))... ( -30.60) >consensus GACAGUAUGUGUUUAUGUUUGCUGAUUUGCUGCUUGUUUGGUUUCGGUUUUCACACAAAUUGCAAUU______GCAAUUGCAGUUGGCUCUGCUGCUA ..((((.(((((....(...(((((...(((........))).)))))...))))))((((((((((........))))))))))......))))... (-20.90 = -20.90 + 0.00)


| Location | 14,698,152 – 14,698,244 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 95.34 |
| Mean single sequence MFE | -17.06 |
| Consensus MFE | -14.80 |
| Energy contribution | -14.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14698152 92 - 20766785 UAGCAGCAGAGCCAACUGCAAUUGC------AAUUGCAAUUUGUGUGAAAACCGAAACCAAACAAGCAGCAAAUCAGCAAACAUAAACACAUACUGUC .....((.((.....((((..((((------....))))((((.((....)))))).........))))....)).)).................... ( -15.20) >DroSec_CAF1 3664 92 - 1 UAGCAGCCGAGCCAACUGCAAUUGC------AAUUGCAAUUUGUGUGAGAACCGAAACCAAACAAGCAGCAAAUCAGCAAACAUAAACACAUACUGUC .....((.((.....((((..((((------....))))((((.((....)))))).........))))....)).)).................... ( -14.40) >DroSim_CAF1 6008 92 - 1 UAGCAGCAGAGCCAACUGCAAUUGC------AAUUGCAAUUUGUGUGAAAACCGAAACCAAACAAGCAGCAAAUCAGCAAACAUAAACACAUACUGUC .....((.((.....((((..((((------....))))((((.((....)))))).........))))....)).)).................... ( -15.20) >DroEre_CAF1 3504 92 - 1 UAGCAGCAGAGCAAACUGCAAUUGC------AAUUGCAAUUUGUGUGAAAACCGAAACCAAACAAGCAGCAAAUCAGCAAACAUAAACACACACUGUC .....((((.((((..(((....))------).))))....((((((......(....).........((......)).........)))))))))). ( -17.10) >DroYak_CAF1 3621 98 - 1 UAGCAGCAGAGCAAACUGCAAUUGCAAUUGCAAUUGCAAUUUGUGUGAAAACCGAAACCAAACAAGCAGCAAAUCAGCAAACAUAAACACACACUGUC .....((((.......(((((((((....)))))))))...((((((......(....).........((......)).........)))))))))). ( -23.40) >consensus UAGCAGCAGAGCCAACUGCAAUUGC______AAUUGCAAUUUGUGUGAAAACCGAAACCAAACAAGCAGCAAAUCAGCAAACAUAAACACAUACUGUC ..((((....((....(((((((........))))))).((((.((....)))))).........)).((......))...............)))). (-14.80 = -14.80 + -0.00)

| Location | 14,698,182 – 14,698,279 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -18.54 |
| Energy contribution | -18.74 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14698182 97 + 20766785 UGCUUGUUUGGUUUCGGUUUUCACACAAAUUGCAAUU------GCAAUUGCAGUUGGCUCUGCUGCUAAACAUAAAGCUCAGUUCCAUGACGCCGGCC-----CCCCU .........((..(((((..(((....(((((((((.------...)))))))))((..(((..(((........))).)))..)).))).)))))..-----))... ( -25.20) >DroSec_CAF1 3694 102 + 1 UGCUUGUUUGGUUUCGGUUCUCACACAAAUUGCAAUU------GCAAUUGCAGUUGGCUCGGCUGCUAAACAUAAAGCUGAGUUCCAUGCCUCACCACCCCCCACCCC ........((((...(((..............(((((------((....)))))))((((((((...........)))))))).....)))..))))........... ( -24.80) >DroSim_CAF1 6038 101 + 1 UGCUUGUUUGGUUUCGGUUUUCACACAAAUUGCAAUU------GCAAUUGCAGUUGGCUCUGCUGCUAAACAUAAAGCUCAGUUCCAUGCCUCCCCCCCUUC-CCCCU .(((((((((((..(((..(..((...((((((....------))))))...))..)..)))..))))))))...)))........................-..... ( -20.00) >DroEre_CAF1 3534 93 + 1 UGCUUGUUUGGUUUCGGUUUUCACACAAAUUGCAAUU------GCAAUUGCAGUUUGCUCUGCUGCUAAACAUAAAGCUCAGUUCCAUAACCCC---------UCCCA .(((((((((((..(((........(((((((((((.------...)))))))))))..)))..))))))))...)))................---------..... ( -22.00) >DroYak_CAF1 3651 98 + 1 UGCUUGUUUGGUUUCGGUUUUCACACAAAUUGCAAUUGCAAUUGCAAUUGCAGUUUGCUCUGCUGCUAAACAUAAAGCUCAGUUCCAUAACCUC---------CCCC- .(((((((((((..(((........((((((((((((((....))))))))))))))..)))..))))))))...)))................---------....- ( -29.90) >consensus UGCUUGUUUGGUUUCGGUUUUCACACAAAUUGCAAUU______GCAAUUGCAGUUGGCUCUGCUGCUAAACAUAAAGCUCAGUUCCAUGACCCC___C_____CCCCU .(((((((((((..(((..........((((((((((........))))))))))....)))..))))))))...))).............................. (-18.54 = -18.74 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:37 2006