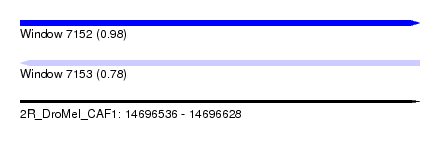
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,696,536 – 14,696,628 |
| Length | 92 |
| Max. P | 0.975598 |

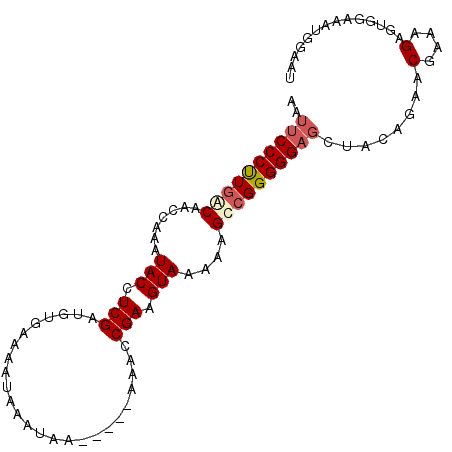
| Location | 14,696,536 – 14,696,628 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 87.39 |
| Mean single sequence MFE | -19.45 |
| Consensus MFE | -16.84 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14696536 92 + 20766785 AUGCAAUUUACACUCUUUCGUU--------CCCCCUGCUUUUUACUUCGGUUUUUUUUUUAUUUAUUUUCACACCGAGGUAUUUGGUUGUCAAGGGAAUU ...................(((--------(((...(((...(((((((((.....................)))))))))...)))......)))))). ( -19.20) >DroSec_CAF1 1980 100 + 1 AUGCCAUUUACACUCUUUCGUUCUGUAGCUCCCCCGGCUUUUUACUUCGGUUUUUUUUUUAUUUAUUUUCACAUCGAGGUAUUUGGUUGUCAAGGGAAUU ..((.((..((........))...)).))((((.(((((...(((((((((.....................)))))))))...)))))....))))... ( -18.00) >DroEre_CAF1 1921 95 + 1 AUUCAAUUUCCACUCUUUCGUGCUGUAGCUCCCCCGACUUUUUACUUCGGUUU-----UUAUUUAUUUUCACAUCGAGGUAUUUGGUUGCCAGGGGAAUU ..........(((......))).......((((((((((...(((((((((..-----..............)))))))))...)))))...)))))... ( -22.49) >DroYak_CAF1 1987 95 + 1 AUUCCAUUUCCACUCUUUCGUUCUGUAGCUCCCCCGACUUUUUACUUCGGUUU-----UUAUUUAUUUUCACAUCGAGGUAUUUGGUUGUCAAGGGAAUU .............................((((.(((((...(((((((((..-----..............)))))))))...)))))....))))... ( -18.09) >consensus AUGCAAUUUACACUCUUUCGUUCUGUAGCUCCCCCGACUUUUUACUUCGGUUU_____UUAUUUAUUUUCACAUCGAGGUAUUUGGUUGUCAAGGGAAUU .............................((((.(((((...(((((((((.....................)))))))))...)))))....))))... (-16.84 = -16.90 + 0.06)

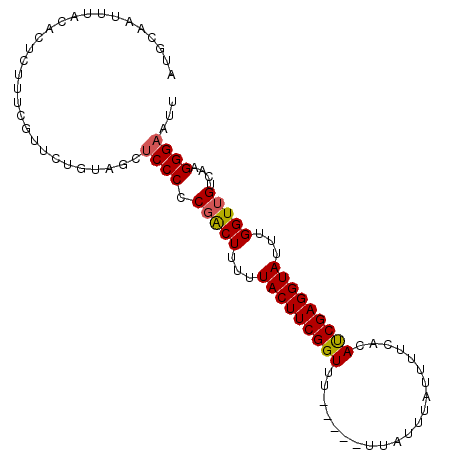
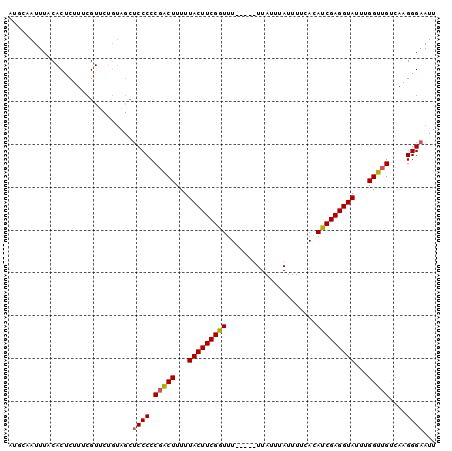
| Location | 14,696,536 – 14,696,628 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 87.39 |
| Mean single sequence MFE | -17.78 |
| Consensus MFE | -12.41 |
| Energy contribution | -13.41 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14696536 92 - 20766785 AAUUCCCUUGACAACCAAAUACCUCGGUGUGAAAAUAAAUAAAAAAAAAACCGAAGUAAAAAGCAGGGGG--------AACGAAAGAGUGUAAAUUGCAU ..((((((((.........(((.(((((.....................))))).))).....)))))))--------).(....).((((.....)))) ( -19.04) >DroSec_CAF1 1980 100 - 1 AAUUCCCUUGACAACCAAAUACCUCGAUGUGAAAAUAAAUAAAAAAAAAACCGAAGUAAAAAGCCGGGGGAGCUACAGAACGAAAGAGUGUAAAUGGCAU ...(((((((.........(((.(((.........................))).)))......)))))))((((....((......)).....)))).. ( -13.77) >DroEre_CAF1 1921 95 - 1 AAUUCCCCUGGCAACCAAAUACCUCGAUGUGAAAAUAAAUAA-----AAACCGAAGUAAAAAGUCGGGGGAGCUACAGCACGAAAGAGUGGAAAUUGAAU ..((((((((((.......(((.(((................-----....))).)))....))))))))))((((....(....).))))......... ( -20.25) >DroYak_CAF1 1987 95 - 1 AAUUCCCUUGACAACCAAAUACCUCGAUGUGAAAAUAAAUAA-----AAACCGAAGUAAAAAGUCGGGGGAGCUACAGAACGAAAGAGUGGAAAUGGAAU ..((((((((((.......(((.(((................-----....))).)))....))))))))))((((....(....).))))......... ( -18.05) >consensus AAUUCCCUUGACAACCAAAUACCUCGAUGUGAAAAUAAAUAA_____AAACCGAAGUAAAAAGCCGGGGGAGCUACAGAACGAAAGAGUGGAAAUGGAAU ..((((((((((.......(((.(((.........................))).)))....))))))))))........(....).............. (-12.41 = -13.41 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:35 2006