| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
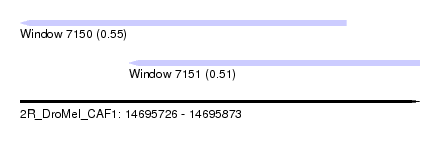
| Location | 14,695,726 – 14,695,873 |
| Length | 147 |
| Max. P | 0.545334 |

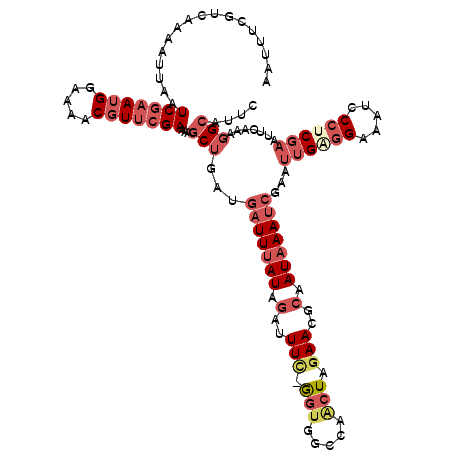
| Location | 14,695,726 – 14,695,846 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -26.59 |
| Consensus MFE | -14.60 |
| Energy contribution | -16.97 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14695726 120 - 20766785 AAUUUCGUCAAAAUUAAUCGAAUGGAAAACGUUCGAAAAGCUGAUGAUUUAUAGAUUUUGGGUUGCCAACUAGAAUGCAAUAAAUCGAAUUGGGGAAAUCCCUCGAAUUCAAAGGCAUUC .................(((((((.....)))))))...(((...(((((((.((((((((........))))))).).)))))))(((((((((....))))..)))))...))).... ( -31.00) >DroSec_CAF1 1094 119 - 1 AAUUUCGUCAAAGUUAAUCGAAUGGAAAACGUUCGAAAAGCUGAUGAUUUAUAGAUUUC-GGUGGCCAACUAGAACGCAAUAAAUCGAAUUGAGGAAAUCCAUCGAAUUCAAAGGCAUUC ......(((..((((..(((((((.....)))))))..))))...(((((((.(..(((-(((.....))).)))..).)))))))(((((((((....)).)).)))))...))).... ( -27.50) >DroSim_CAF1 3499 119 - 1 AAUUUCGUCAAAGUUAAUCGAAUGGAAAACGUUCGAAAAGCUGAUGAUUUAUAGAUUUC-GGUGGCCAACUAGAACGCAAUAAAUCGAAUUGAGGAAAUCCCUCGAAUUCAAAGGCAUUC ......(((..((((..(((((((.....)))))))..))))...(((((((.(..(((-(((.....))).)))..).)))))))(((((((((.....)))).)))))...))).... ( -30.70) >DroEre_CAF1 1014 117 - 1 --UUUCGUCAAAAUUAAUCGAAUGGAAAGCGUUCGAAAAGCUGAUGAUUUAUAGAUUUC-AGUGGCCAACUAGAACGCAAUAAAUCCAAUUGGGGAAAUCCCUCGAAUGCAAAGGCAUUC --................(((.((((..((((((.....(((((.((((....))))))-))).........))))))......)))).)))(((....)))..((((((....)))))) ( -28.04) >DroYak_CAF1 1045 119 - 1 AAUUUCGUCAAAAUUAAUCGAAUGGAAAGCGUUCGAAAAGCUGAUGAUUUAUAGAUUUC-GGUGGCCAGCUAGAACGCAAUAAAUCCAAUUGAGGAAAUCCCCCGAAUGCAAAGGAAUUC ((((((...........((((.((((..((((((....(((((.(.((..(......).-.)).).))))).))))))......)))).))))((....))............)))))). ( -24.00) >DroAna_CAF1 437 96 - 1 AAUC----------UAAUCACA-GGAAGACCU--GAAAAGCACACUUUUCAU------C-AAAAACCAACU----UGCCAUAAAUCGAAAAGCGGAAACCCCUCGAAAUCAAAGGCUUCC ....----------........-(((((.(((--((((((....))))))..------.-...........----.........((((...(.(....))..))))......)))))))) ( -18.30) >consensus AAUUUCGUCAAAAUUAAUCGAAUGGAAAACGUUCGAAAAGCUGAUGAUUUAUAGAUUUC_GGUGGCCAACUAGAACGCAAUAAAUCGAAUUGAGGAAAUCCCUCGAAUUCAAAGGCAUUC .................(((((((.....)))))))...(((...(((((((.(..(((.(((.....))).)))..).)))))))...((((((.....)))))).......))).... (-14.60 = -16.97 + 2.37)


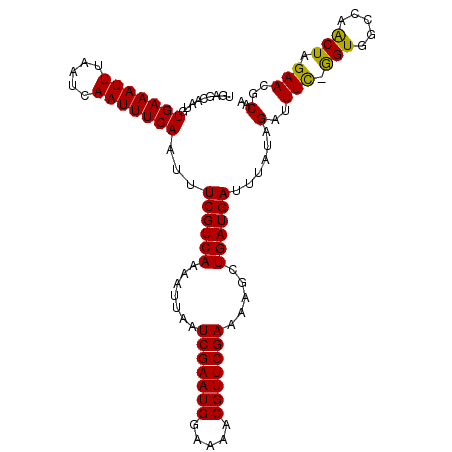
| Location | 14,695,766 – 14,695,873 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -21.49 |
| Consensus MFE | -15.91 |
| Energy contribution | -15.78 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14695766 107 - 20766785 UGAGCAAUGUGAAAUUUAAUCAAUUUCAAUUUCGUCAAAAUUAAUCGAAUGGAAAACGUUCGAAAAGCUGAUGAUUUAUAGAUUUUGGGUUGCCAACUAGAAUGCAA ((.(((((.(((((((((.............((((((.......(((((((.....))))))).....))))))....))))))))).)))))))............ ( -22.73) >DroSec_CAF1 1134 106 - 1 UGAGCAAUGUGAAAUUUAAUCAAUUUCAAUUUCGUCAAAGUUAAUCGAAUGGAAAACGUUCGAAAAGCUGAUGAUUUAUAGAUUUC-GGUGGCCAACUAGAACGCAA ...((....(((((((.....)))))))...((((((..(((..(((((((.....)))))))..)))))))))........(((.-.((.....))..))).)).. ( -21.90) >DroSim_CAF1 3539 106 - 1 UGAGCAAUGUGAAAUUUAAUCAAUUUCAAUUUCGUCAAAGUUAAUCGAAUGGAAAACGUUCGAAAAGCUGAUGAUUUAUAGAUUUC-GGUGGCCAACUAGAACGCAA ...((....(((((((.....)))))))...((((((..(((..(((((((.....)))))))..)))))))))........(((.-.((.....))..))).)).. ( -21.90) >DroEre_CAF1 1054 84 - 1 UGAGAAA----------------------UUUCGUCAAAAUUAAUCGAAUGGAAAGCGUUCGAAAAGCUGAUGAUUUAUAGAUUUC-AGUGGCCAACUAGAACGCAA ...((((----------------------((((((((.......(((((((.....))))))).....))))))......))))))-.(((..(.....)..))).. ( -16.90) >DroYak_CAF1 1085 106 - 1 UGAGAAAUGUGAAAUUUAAUCAAUUUCAAUUUCGUCAAAAUUAAUCGAAUGGAAAGCGUUCGAAAAGCUGAUGAUUUAUAGAUUUC-GGUGGCCAGCUAGAACGCAA ((((((((.(((((((.....)))))))))))).)))..................((((((....(((((.(.((..(......).-.)).).))))).)))))).. ( -24.00) >consensus UGAGCAAUGUGAAAUUUAAUCAAUUUCAAUUUCGUCAAAAUUAAUCGAAUGGAAAACGUUCGAAAAGCUGAUGAUUUAUAGAUUUC_GGUGGCCAACUAGAACGCAA .........(((((((.....)))))))...((((((.......(((((((.....))))))).....))))))......(..(((.(((.....))).)))..).. (-15.91 = -15.78 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:33 2006