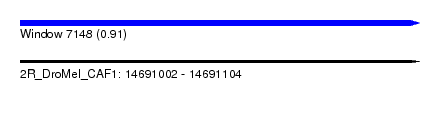
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,691,002 – 14,691,104 |
| Length | 102 |
| Max. P | 0.909300 |

| Location | 14,691,002 – 14,691,104 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -13.00 |
| Consensus MFE | -9.75 |
| Energy contribution | -9.50 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
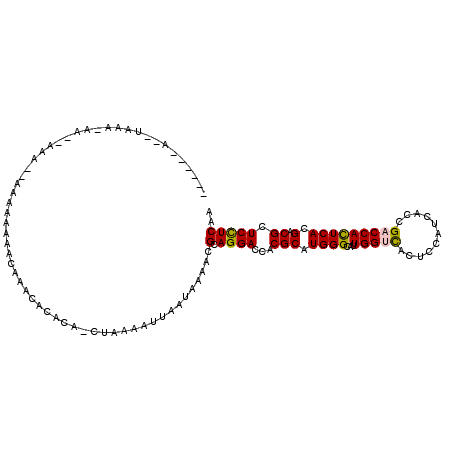
>2R_DroMel_CAF1 14691002 102 + 20766785 ------A--UAAA-AA--AAA--AACAAAAACAAACACACA-CUAAAAUUAAUAAAACGCAGGACCACGCAUGGGCAUUGGUCACUCCAUCACCGACCACUCACGUCGCUCCUCAA ------.--....-..--...--..................-................(.(((((.(((..((((...(((((...........)))))))))))).).))))).. ( -13.90) >DroVir_CAF1 20452 114 + 1 AUAUUCAAUUCAAUCA--ACAAAAUAUAUAACAACUUCACAACAAAAAUCAUGCAAAUGAAGGACCCCGCAUGGGAAUUGGCUCAUCCAUCACCGACCACUCACGACGCUCUUCAA ................--.......................................(((((..((((....)))...(((.((..........)))))........)..))))). ( -11.00) >DroSim_CAF1 18356 101 + 1 ------A--UAAA-AU--AA---AAAAAAAACAAACACACA-CUAAAAUUAAUAAAACGCAGGACCACGCAUGGGCAUUGGUCACUCCAUCACCGACCACUCACGCCGCUCCUCAA ------.--....-..--..---..................-................(.((((.(..((.((((...(((((...........))))))))).)).).))))).. ( -14.30) >DroEre_CAF1 20754 106 + 1 ------A--UAAA-AAUUAAAAAACAAAAAACAAACACACA-CUAAAAUUAAUAAAACGCAGGACCACGCAUGGGCAUUGGUCACUCCAUCACCGACCACUCACGACGCUCCUCAA ------.--....-...........................-................(.((((...(((.((((...(((((...........))))))))).).)).))))).. ( -12.80) >DroYak_CAF1 21271 104 + 1 ------A--UAAA-AA--AAAAAAUAAAACAAAAACACACA-CUAAAUUUAAUAAAACGCAGGACCACGCAUGGGCAUUGGUCACUCCAUCACCGACCACUCACGCCGCUCCUCAA ------.--....-..--.......................-................(.((((.(..((.((((...(((((...........))))))))).)).).))))).. ( -14.30) >DroPer_CAF1 28438 92 + 1 ------U--CGAA-CA--CAC--UAUUAAAACGAA---ACA-CUAAAUU-------ACGCAGGACCCCGCAUGGGCAUUGGCUCCGCCAUCACCGACCAUUCACGACGCUCUUCAA ------.--....-..--...--.........(((---...-.......-------..((.(....).))..((((..((((...))))....((........))..))))))).. ( -11.70) >consensus ______A__UAAA_AA__AAA__AAAAAAAACAAACACACA_CUAAAAUUAAUAAAACGCAGGACCACGCAUGGGCAUUGGUCACUCCAUCACCGACCACUCACGACGCUCCUCAA ..........................................................(.((((...(((.((((...(((((...........))))))))).).)).))))).. ( -9.75 = -9.50 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:30 2006