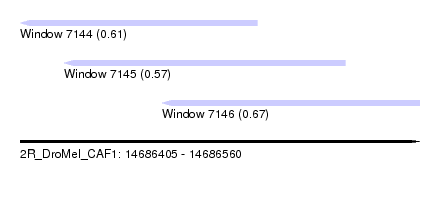
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,686,405 – 14,686,560 |
| Length | 155 |
| Max. P | 0.668335 |

| Location | 14,686,405 – 14,686,497 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 82.37 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -20.42 |
| Energy contribution | -22.15 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14686405 92 - 20766785 AAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUUCCCAGUUUGCCAGUUGGACGGCGAAGGGGAAA-GAGCUGGAGCCGAAGCAGCU-GAA ................((((((.(((((.(((...(((((..((((((........)))))).))))).-..))).).))))....))))-)). ( -33.60) >DroSec_CAF1 17491 92 - 1 AAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUUGCCAGUUCGCCAGUUGGACGGCGAAAGGGAAA-GAGCUGGAGCCGAAGCAGCU-GAA ................((((((.(((((.(((......((..((((((........))))))..))...-..))).).))))....))))-)). ( -30.50) >DroSim_CAF1 12591 92 - 1 AAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUUCCCAGUUCGCCAGUUGGCCGGCGAAGGGGAAA-GAGCUGGAGCCGAAGCAGCU-GAA ................((((((.(((((.(((...(((((..((((((.(....).)))))).))))).-..))).).))))....))))-)). ( -36.00) >DroEre_CAF1 16263 92 - 1 AAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUUCCCAGUUCGCCAGUUGGCUGGAGGAGGGGAAAAGCGCUGGGGCC-AAGCAGCU-GAA ................((((((..((((.((((..(((((..(((.((((....)))).))).)))))...)))).).)))-....))))-)). ( -34.40) >DroYak_CAF1 16643 80 - 1 AAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUUCCCAGUUCGCCGGCUGGCUGGAGAAGGGGAAAA-------------AGCAGCU-GAA .....((.........(((((....)))))((...(((((..(((.((((....)))).))).)))))..-------------.)).)).-... ( -27.00) >DroAna_CAF1 14145 84 - 1 AAAACGCGUUUUGCCAUCAGUUACGGCUGAGCGAGUUCCCAGCUUG------GAGCUGCGAGGGGCUCG-GAGC---AGCGGAGGCAGCUCUAA .......((((.(((.........))).))))(((((((((((...------..))))...)))))))(-((((---.((....)).))))).. ( -30.70) >consensus AAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUUCCCAGUUCGCCAGUUGGCCGGCGAAGGGGAAA_GAGCUGGAGCCGAAGCAGCU_GAA .....((((((((...(((((....)))))((...(((((..((((((........)))))).)))))....))......)))))).))..... (-20.42 = -22.15 + 1.73)

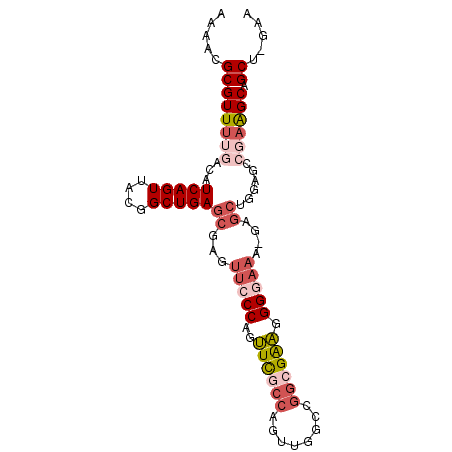
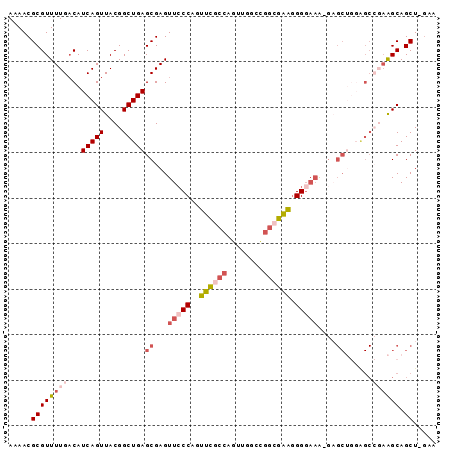
| Location | 14,686,422 – 14,686,531 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -23.96 |
| Energy contribution | -25.13 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14686422 109 - 20766785 AGCUAAAUACUACACGGUAU----AGAAAACUGAAAACAAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUUCCCAGUUUGCCAGUUGGACGGCGAAGGGGAAA-GAGCUG ((((..(((((....)))))----....((((((...(((((....)))))...))))))..)))).(((...(((((..((((((........)))))).))))).-..))). ( -33.90) >DroSec_CAF1 17508 109 - 1 AGCUAAAUACUACACGGUAU----AGAAAACUGAAAACAAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUUGCCAGUUCGCCAGUUGGACGGCGAAAGGGAAA-GAGCUG ((((..(((((....)))))----....((((((...(((((....)))))...))))))..)))).(((......((..((((((........))))))..))...-..))). ( -30.80) >DroSim_CAF1 12608 109 - 1 AGCUAAAUACUACACGGUAU----AGAAAACUGAAAACAAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUUCCCAGUUCGCCAGUUGGCCGGCGAAGGGGAAA-GAGCUG ((((..(((((....)))))----....((((((...(((((....)))))...))))))..)))).(((...(((((..((((((.(....).)))))).))))).-..))). ( -36.30) >DroEre_CAF1 16279 110 - 1 AGCUAAAUACUACACGGUAU----AGAAAACUGAAAACAAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUUCCCAGUUCGCCAGUUGGCUGGAGGAGGGGAAAAGCGCUG ((((..(((((....)))))----....((((((...(((((....)))))...))))))..)))).((((..(((((..(((.((((....)))).))).)))))...)))). ( -36.80) >DroYak_CAF1 16653 104 - 1 AGCUAAAUACUACACGGUAU----AGAAAACUGAAAACAAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUUCCCAGUUCGCCGGCUGGCUGGAGAAGGGGAAAA------ ((((..(((((....)))))----....((((((...(((((....)))))...))))))..)))).......(((((..(((.((((....)))).))).)))))..------ ( -29.60) >DroAna_CAF1 14162 102 - 1 CGCCA---GCUAGACGGCGUGGAGAAAAAGUUGAAAACAAAACGCGUUUUGCCAUCAGUUACGGCUGAGCGAGUUCCCAGCUUG------GAGCUGCGAGGGGCUCG-GAGC-- .((((---(((.(((...((((..(((..(((........)))...)))..))))..)))..))))).))(((((((((((...------..))))...))))))).-....-- ( -34.30) >consensus AGCUAAAUACUACACGGUAU____AGAAAACUGAAAACAAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUUCCCAGUUCGCCAGUUGGCCGGCGAAGGGGAAA_GAGCUG ((((..(((((....)))))........((((((...(((((....)))))...))))))..)))).(((...(((((..((((((........)))))).)))))....))). (-23.96 = -25.13 + 1.17)

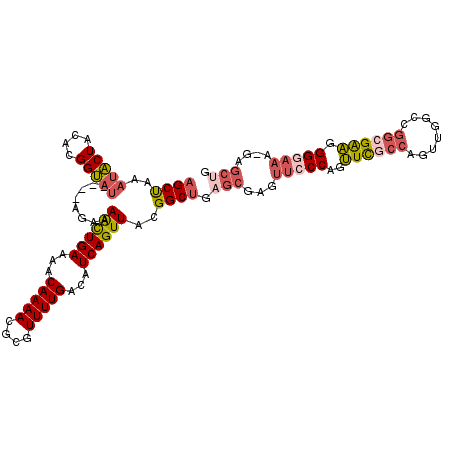
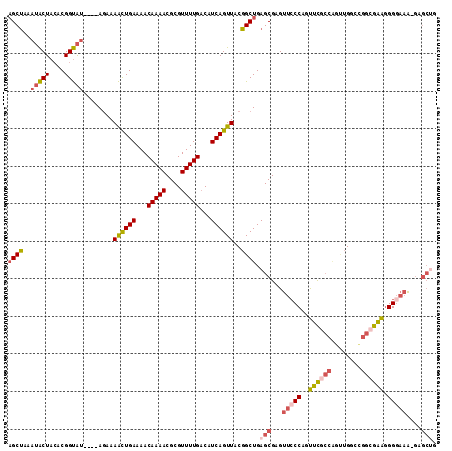
| Location | 14,686,460 – 14,686,560 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 85.65 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -19.31 |
| Energy contribution | -19.15 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
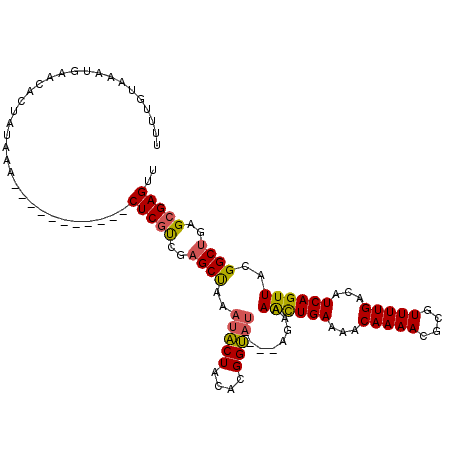
>2R_DroMel_CAF1 14686460 100 - 20766785 UUUUGUAAAUCAUCACUAUCAA-----------CUCGUCGAGCUAAAUACUACACGGUAU----AGAAAACUGAAAACAAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUU ....................((-----------(((((..((((..(((((....)))))----....((((((...(((((....)))))...))))))..))))..))))))) ( -24.20) >DroSec_CAF1 17546 100 - 1 UUUUGUAAAUGAACACUAUCAA-----------CUCGUCGAGCUAAAUACUACACGGUAU----AGAAAACUGAAAACAAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUU ....................((-----------(((((..((((..(((((....)))))----....((((((...(((((....)))))...))))))..))))..))))))) ( -24.20) >DroSim_CAF1 12646 100 - 1 UUUUGUAAAUGAACACUAUCAA-----------CUCGUCGAGCUAAAUACUACACGGUAU----AGAAAACUGAAAACAAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUU ....................((-----------(((((..((((..(((((....)))))----....((((((...(((((....)))))...))))))..))))..))))))) ( -24.20) >DroEre_CAF1 16318 100 - 1 UUUUGUAAACGAACACUAUAAG-----------CUCGUCGAGCUAAAUACUACACGGUAU----AGAAAACUGAAAACAAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUU .((((....)))).......((-----------(((((..((((..(((((....)))))----....((((((...(((((....)))))...))))))..))))..))))))) ( -24.30) >DroYak_CAF1 16686 100 - 1 UUUUGUAACUAAACACUAUAAA-----------CUCGUCGAGCUAAAUACUACACGGUAU----AGAAAACUGAAAACAAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUU ....................((-----------(((((..((((..(((((....)))))----....((((((...(((((....)))))...))))))..))))..))))))) ( -24.20) >DroAna_CAF1 14192 110 - 1 UUUAUUAAACAAA--UUAUAAGCCCAACAUUCACUCUCCUCGCCA---GCUAGACGGCGUGGAGAAAAAGUUGAAAACAAAACGCGUUUUGCCAUCAGUUACGGCUGAGCGAGUU .............--.........((((......(((((.((((.---.......)))).)))))....)))).......(((.(((((.(((.........))).))))).))) ( -28.40) >consensus UUUUGUAAAUGAACACUAUAAA___________CUCGUCGAGCUAAAUACUACACGGUAU____AGAAAACUGAAAACAAAACGCGUUUUGACAUCAGUUACGGCUGAGCGAGUU .................................(((((..((((..(((((....)))))........((((((...(((((....)))))...))))))..))))..))))).. (-19.31 = -19.15 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:28 2006