| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,678,862 – 14,678,992 |
| Length | 130 |
| Max. P | 0.894829 |

| Location | 14,678,862 – 14,678,967 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.59 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -15.53 |
| Energy contribution | -16.60 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14678862 105 + 20766785 GAUCAAUAAAAAAUAUAU------------U-UUUUAGAAACCAUGUUGCGCAACAUUUGCAGGGAGCAACAAGUGCCUAAACAUGGCUAUCGAUACCAAGACGACAAAUAUCGAUAA ......(((((((....)------------)-)))))....(((((((..(((....((((.....))))....)))...))))))).((((((((.............)))))))). ( -22.72) >DroSim_CAF1 5040 111 + 1 ---UGAUAAGAAAUAUAUU---GACCGAAUU-UUUUACAAACCAUGUUGCGCAACAUUUGCAGGGAGCAACAAGUGCCUAAACAUGGCUAUCGAUACCAAGACGACAAAUAUCGAUAA ---((.((((((((.....---......)))-)))))))..(((((((..(((....((((.....))))....)))...))))))).((((((((.............)))))))). ( -23.32) >DroEre_CAF1 8491 111 + 1 G----AGAAAAAAUUUAGUCUUGAUCGAAUUUC---AGCAACCAUGUUGCGCAACAUUUGCAUGGAGCAACAAGUGCCUAAACAUUGUUCUCGAUACCAAUAGGACAAGUAUCGAUAA (----((((....(((((.((((..........---.(((((...)))))((..(((....)))..))..))))...))))).....)))))(((((...........)))))..... ( -23.20) >DroYak_CAF1 8718 114 + 1 G----AUAAUGAAAUUAAUUUUUUUCUAAUUUUUGUAGAAACCAUGUUGCGCAACAUUUUUAUGGAGCAACAAGUGCCUAAAAACGGCUAUCGAUACCAGACCGACAAAUAUCGAUAA .----.................((((((.......))))))...((((((....(((....)))..))))))...(((.......)))((((((((.............)))))))). ( -20.22) >consensus G____AUAAAAAAUAUAUU___GAUCGAAUU_UUUUAGAAACCAUGUUGCGCAACAUUUGCAGGGAGCAACAAGUGCCUAAACAUGGCUAUCGAUACCAAGACGACAAAUAUCGAUAA .........................................(((((((..(((....((((.....))))....)))...))))))).((((((((.............)))))))). (-15.53 = -16.60 + 1.06)


| Location | 14,678,862 – 14,678,967 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.59 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -20.44 |
| Energy contribution | -21.13 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14678862 105 - 20766785 UUAUCGAUAUUUGUCGUCUUGGUAUCGAUAGCCAUGUUUAGGCACUUGUUGCUCCCUGCAAAUGUUGCGCAACAUGGUUUCUAAAA-A------------AUAUAUUUUUUAUUGAUC .((((((((((.........))))))))))((((((((...(((....((((.....))))....)))..))))))))...(((((-(------------(....)))))))...... ( -26.30) >DroSim_CAF1 5040 111 - 1 UUAUCGAUAUUUGUCGUCUUGGUAUCGAUAGCCAUGUUUAGGCACUUGUUGCUCCCUGCAAAUGUUGCGCAACAUGGUUUGUAAAA-AAUUCGGUC---AAUAUAUUUCUUAUCA--- .((((((((((.........))))))))))((((((((...(((....((((.....))))....)))..))))))))........-.........---................--- ( -25.60) >DroEre_CAF1 8491 111 - 1 UUAUCGAUACUUGUCCUAUUGGUAUCGAGAACAAUGUUUAGGCACUUGUUGCUCCAUGCAAAUGUUGCGCAACAUGGUUGCU---GAAAUUCGAUCAAGACUAAAUUUUUUCU----C ...((((((((.........))))))))(((....(((((((((....((((.....))))....)))(((((...))))).---...............))))))...))).----. ( -25.80) >DroYak_CAF1 8718 114 - 1 UUAUCGAUAUUUGUCGGUCUGGUAUCGAUAGCCGUUUUUAGGCACUUGUUGCUCCAUAAAAAUGUUGCGCAACAUGGUUUCUACAAAAAUUAGAAAAAAAUUAAUUUCAUUAU----C .(((((((((..(.....)..)))))))))(((.......))).(.((((((.(.(((....))).).)))))).).((((((.......)))))).................----. ( -24.30) >consensus UUAUCGAUAUUUGUCGUCUUGGUAUCGAUAGCCAUGUUUAGGCACUUGUUGCUCCAUGCAAAUGUUGCGCAACAUGGUUUCUAAAA_AAUUCGAUC___AAUAAAUUUUUUAU____C .((((((((((.........))))))))))((((((((...(((....((((.....))))....)))..))))))))........................................ (-20.44 = -21.13 + 0.69)

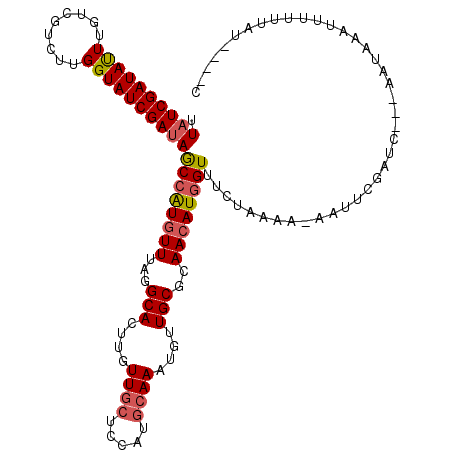
| Location | 14,678,887 – 14,678,992 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.25 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -15.66 |
| Energy contribution | -16.70 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14678887 105 + 20766785 AAACCAUGUUGCGCAACAUUUGCAGGGAGCAACAAGUGCCUAAACAUGGCUAUCGAUACCAAGACGACAAAUAUCGAUAAUU----------UUUCUUCUACUCUUUUAACAUUA ...(((((((..(((....((((.....))))....)))...))))))).((((((((.............))))))))...----------....................... ( -21.62) >DroSec_CAF1 9996 115 + 1 AAACCAUGUUGCACAACAUUUGCAGGGAGCAACAAGUGCCUAAACAUGGCUAUCGAUACCAAGACUACAAAUAUCGAUAUAUUCCCAUGCUCUCUCUCCUACUCUUUUAACAUUA .....((((((((.......)))((((((((......(((.......)))((((((((.............))))))))........)))))))).............))))).. ( -24.62) >DroSim_CAF1 5071 115 + 1 AAACCAUGUUGCGCAACAUUUGCAGGGAGCAACAAGUGCCUAAACAUGGCUAUCGAUACCAAGACGACAAAUAUCGAUAAUUUCCCACGUACUCUCUGCUACUCUGUUAACAUUA .....((((((.(((......((((((((..((....(((.......)))((((((((.............)))))))).........)).)))))))).....))))))))).. ( -27.52) >DroEre_CAF1 8522 114 + 1 CAACCAUGUUGCGCAACAUUUGCAUGGAGCAACAAGUGCCUAAACAUUGUUCUCGAUACCAAUAGGACAAGUAUCGAUAACCGCCCAUGUUCUCUC-UUUACUCUUUUAACAUUC .....(((((....)))))..((((((.(((((((((......)).))))).(((((((...........))))))).....))))))))......-.................. ( -21.90) >DroYak_CAF1 8752 114 + 1 AAACCAUGUUGCGCAACAUUUUUAUGGAGCAACAAGUGCCUAAAAACGGCUAUCGAUACCAGACCGACAAAUAUCGAUAACACCCCA-GUUCUCUCCUUUACGCUUUUAACAUUC .....((((((.((..(((....)))(((.(((....(((.......)))((((((((.............))))))))........-))))))........))...)))))).. ( -18.62) >consensus AAACCAUGUUGCGCAACAUUUGCAGGGAGCAACAAGUGCCUAAACAUGGCUAUCGAUACCAAGACGACAAAUAUCGAUAACUUCCCA_GUUCUCUCUUCUACUCUUUUAACAUUA ...(((((((..(((....((((.....))))....)))...))))))).((((((((.............)))))))).................................... (-15.66 = -16.70 + 1.04)

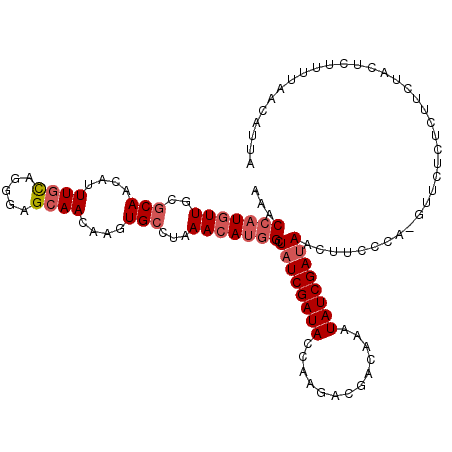
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:23 2006