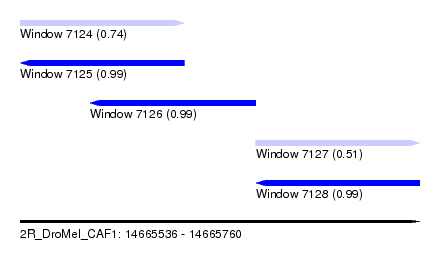
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,665,536 – 14,665,760 |
| Length | 224 |
| Max. P | 0.994985 |

| Location | 14,665,536 – 14,665,628 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.51 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -21.26 |
| Energy contribution | -21.07 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14665536 92 + 20766785 UUUAUUCUGGAUAAUUGCCCAGGGGCUUAAUUUUCGGUGGUGUGUGCUUUU---------------UUUUUGGCAGGGGGAAGCGGCACAUAUGCGCCUACCA------------CCCA .........((.(((((((....)))..)))).))(((((((...((((((---------------(((......)))))))))(((.(....).))))))))------------)).. ( -28.40) >DroSim_CAF1 20127 98 + 1 UUUAUUCUGGAUAAUUGCCCAGGGGCUUAAUUUUCGGUGGUGUGUGCUUUUCUUUU---------UUUUUGGACAGGGGGAAGCGGCACAUAUGCGCCUGCCA------------CCCA .........((.(((((((....)))..)))).))((((((....((((..((((.---------((....)).))))..))))(((.(....).))).))))------------)).. ( -30.70) >DroEre_CAF1 20380 110 + 1 UUUAUUCUGGAUAAUUGCCCAGGGGCUUAAUUUUCGGUGGUGUGUGCUUUU---------UUUUUGUUUUGGACAGGGGGAAGCGGCACAUAUGCGCCUACCACCCACCAUACCACCGA ....((((((........)))))).........(((((((((((.((((((---------((((.(((...)))))))))))))(((.(....).)))..........))))))))))) ( -36.80) >DroYak_CAF1 20438 117 + 1 UUUAUUCUGGAUAAUUGCCCAGGGGCUUAAUUUUCGGUGGUGUGUGUUUUUU--UCUCUUUUUUUUUUUUGGACGGGGGGAAGCGGCACAUAUGCGCCUACCACCCACCACACCACCGA ....((((((........)))))).........(((((((((((.((((((.--.((.(((.........))).))..))))))(((.(....).)))..........))))))))))) ( -38.70) >consensus UUUAUUCUGGAUAAUUGCCCAGGGGCUUAAUUUUCGGUGGUGUGUGCUUUU______________UUUUUGGACAGGGGGAAGCGGCACAUAUGCGCCUACCA____________CCCA .......(((......(((....))).........(((((((((((((....................................))))))))).))))..)))................ (-21.26 = -21.07 + -0.19)

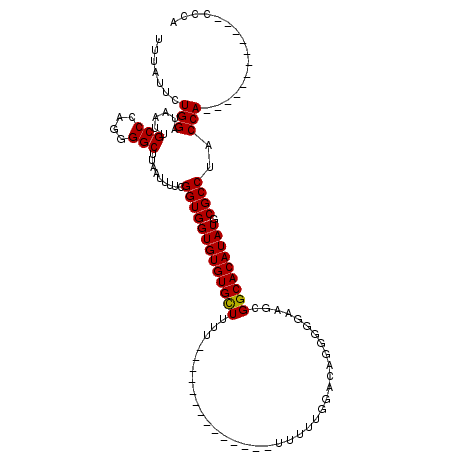
| Location | 14,665,536 – 14,665,628 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.51 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -19.71 |
| Energy contribution | -20.78 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14665536 92 - 20766785 UGGG------------UGGUAGGCGCAUAUGUGCCGCUUCCCCCUGCCAAAAA---------------AAAAGCACACACCACCGAAAAUUAAGCCCCUGGGCAAUUAUCCAGAAUAAA ..((------------((((.(((((....)))))((((..............---------------..))))....)))))).........(((....)))................ ( -26.89) >DroSim_CAF1 20127 98 - 1 UGGG------------UGGCAGGCGCAUAUGUGCCGCUUCCCCCUGUCCAAAAA---------AAAAGAAAAGCACACACCACCGAAAAUUAAGCCCCUGGGCAAUUAUCCAGAAUAAA ..((------------(((..(((((....)))))((((....((.........---------...))..)))).....))))).........(((....)))................ ( -25.00) >DroEre_CAF1 20380 110 - 1 UCGGUGGUAUGGUGGGUGGUAGGCGCAUAUGUGCCGCUUCCCCCUGUCCAAAACAAAAA---------AAAAGCACACACCACCGAAAAUUAAGCCCCUGGGCAAUUAUCCAGAAUAAA ((((((((.(((((((.((..(((((....)))))....)))))(((.....)))....---------....)).)).)))))))).......(((....)))................ ( -37.90) >DroYak_CAF1 20438 117 - 1 UCGGUGGUGUGGUGGGUGGUAGGCGCAUAUGUGCCGCUUCCCCCCGUCCAAAAAAAAAAAAGAGA--AAAAAACACACACCACCGAAAAUUAAGCCCCUGGGCAAUUAUCCAGAAUAAA ((((((((((((((((.((..(((((....)))))....)).))).(((............).))--.....)).))))))))))).......(((....)))................ ( -42.00) >consensus UCGG____________UGGUAGGCGCAUAUGUGCCGCUUCCCCCUGUCCAAAAA______________AAAAGCACACACCACCGAAAAUUAAGCCCCUGGGCAAUUAUCCAGAAUAAA ..((..........((((((.(((((....)))))((........))...............................)))))).........(((....)))......))........ (-19.71 = -20.78 + 1.06)

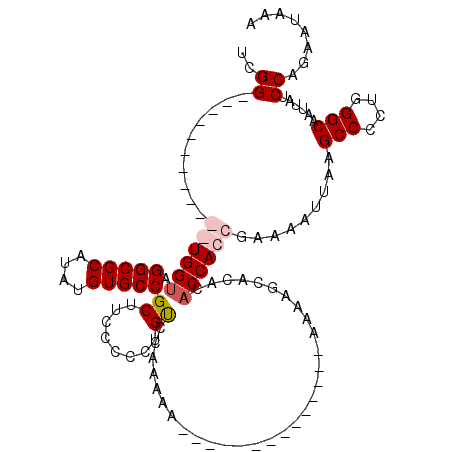
| Location | 14,665,575 – 14,665,668 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.13 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -23.14 |
| Energy contribution | -24.89 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14665575 93 - 20766785 AAAGCCAUUGAAGGGCAACUUCAAGGUGUGGGCGGCAUUGUGGG------------UGGUAGGCGCAUAUGUGCCGCUUCCCCCUGCCAAAAA---------------AAAAGCACACAC ...(((.((((((.....)))))))))(((.((((((..(.(((------------.(((.(((((....)))))))).)))).)))).....---------------....)).))).. ( -33.80) >DroSim_CAF1 20166 99 - 1 AAAGCCAUUGAAGGGCAACUUCAAGGUGUGGGCGGCAAAAUGGG------------UGGCAGGCGCAUAUGUGCCGCUUCCCCCUGUCCAAAAA---------AAAAGAAAAGCACACAC ...(((.((((((.....))))))))).(((((((......(((------------.(((.(((((....)))))))).))).)))))))....---------................. ( -34.90) >DroEre_CAF1 20419 111 - 1 AAAGCCAUUGAAGGGCAACAUCGAGGUGUGGGCGGGAAAAUCGGUGGUAUGGUGGGUGGUAGGCGCAUAUGUGCCGCUUCCCCCUGUCCAAAACAAAAA---------AAAAGCACACAC ...(((.((((..(....).))))))).((((((((...((((......))))(((.(((.(((((....)))))))).))))))))))).........---------............ ( -35.70) >DroYak_CAF1 20477 118 - 1 AAAGCCAUUGAAGGGCAACUUCAAGAAGUAAGCGGGAAAAUCGGUGGUGUGGUGGGUGGUAGGCGCAUAUGUGCCGCUUCCCCCCGUCCAAAAAAAAAAAAGAGA--AAAAAACACACAC ...((((((((((.....))))).........(((.....)))))))).(((((((.((..(((((....)))))....)).)))).)))...............--............. ( -31.70) >consensus AAAGCCAUUGAAGGGCAACUUCAAGGUGUGGGCGGCAAAAUCGG____________UGGUAGGCGCAUAUGUGCCGCUUCCCCCUGUCCAAAAA______________AAAAGCACACAC ...(((.((((((.....))))))))).((((((((.....(((.............(((.(((((....)))))))).))))))))))).............................. (-23.14 = -24.89 + 1.75)

| Location | 14,665,668 – 14,665,760 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 94.23 |
| Mean single sequence MFE | -19.14 |
| Consensus MFE | -16.46 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14665668 92 + 20766785 CUUUUGUUUUGGAAUAUGCGAAAAUUAACUGGUAAAAUGUUGGCCCAUCAACACGGGGGUAACUUCAACAUUUUCCAAUUAAAAAAGUCACC (((((.(((((.......))))).((((.(((.(((((((((((((.((.....)))))).....)))))))))))).)))))))))..... ( -19.20) >DroSec_CAF1 20226 92 + 1 CUUUUGUUUUGGAAUAUGCGAAAAUUAACUGGUAAAAUGUUGGCCCAUCAACACGGGGGUAAAUCCAACAUUUUCCAAUUAAAAAAGUCACC (((((.(((((.......))))).((((.(((.(((((((((((((.((.....))))).....))))))))))))).)))))))))..... ( -20.50) >DroSim_CAF1 20265 92 + 1 CUUUUGUUUUGGAAUAUGCGAAAAUUAACUGGUAAAAUGUUGGCCCAUCAACACGGGGGUAAAUCCAACAUUUUCCAAUUAAAAAAGUCACC (((((.(((((.......))))).((((.(((.(((((((((((((.((.....))))).....))))))))))))).)))))))))..... ( -20.50) >DroEre_CAF1 20530 91 + 1 CUGUUGUUUCGGAGUAUGCGAAAAUUAACUGGUAAAAUGUUGGCCCAUCAACACAGGGGUAACU-CAACAUUUUCCAAUUAAAAAAGUCACC ......(((((.......))))).((((.(((.(((((((((((((..........))))....-)))))))))))).)))).......... ( -19.60) >DroYak_CAF1 20595 91 + 1 CCUUUGUUUGGGAGUAUGCGAAAAUUAACUGGUAAAAUGUUGGCCCAUCAACGCAGGGGUAACU-CUACGUUUUCCAAUUAAAAAAGUCACC (((......))).......((...((((.(((.(((((((..((((..........))))....-..)))))))))).)))).....))... ( -15.90) >consensus CUUUUGUUUUGGAAUAUGCGAAAAUUAACUGGUAAAAUGUUGGCCCAUCAACACGGGGGUAACU_CAACAUUUUCCAAUUAAAAAAGUCACC .((((((..........)))))).((((.(((.(((((((((((((..........)))).....)))))))))))).)))).......... (-16.46 = -16.90 + 0.44)

| Location | 14,665,668 – 14,665,760 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 94.23 |
| Mean single sequence MFE | -24.14 |
| Consensus MFE | -20.12 |
| Energy contribution | -20.48 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.994985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14665668 92 - 20766785 GGUGACUUUUUUAAUUGGAAAAUGUUGAAGUUACCCCCGUGUUGAUGGGCCAACAUUUUACCAGUUAAUUUUCGCAUAUUCCAAAACAAAAG .((((.....(((((((((((((((((........(((((....))))).))))))))).))))))))...))))................. ( -23.30) >DroSec_CAF1 20226 92 - 1 GGUGACUUUUUUAAUUGGAAAAUGUUGGAUUUACCCCCGUGUUGAUGGGCCAACAUUUUACCAGUUAAUUUUCGCAUAUUCCAAAACAAAAG .((((.....((((((((((((((((((.......(((((....))))))))))))))).))))))))...))))................. ( -27.41) >DroSim_CAF1 20265 92 - 1 GGUGACUUUUUUAAUUGGAAAAUGUUGGAUUUACCCCCGUGUUGAUGGGCCAACAUUUUACCAGUUAAUUUUCGCAUAUUCCAAAACAAAAG .((((.....((((((((((((((((((.......(((((....))))))))))))))).))))))))...))))................. ( -27.41) >DroEre_CAF1 20530 91 - 1 GGUGACUUUUUUAAUUGGAAAAUGUUG-AGUUACCCCUGUGUUGAUGGGCCAACAUUUUACCAGUUAAUUUUCGCAUACUCCGAAACAACAG .((((.....(((((((((((((((((-.(....(((.........))))))))))))).))))))))...))))................. ( -22.30) >DroYak_CAF1 20595 91 - 1 GGUGACUUUUUUAAUUGGAAAACGUAG-AGUUACCCCUGCGUUGAUGGGCCAACAUUUUACCAGUUAAUUUUCGCAUACUCCCAAACAAAGG .((((.....((((((((..(((((((-........)))))))((((......))))...))))))))...))))................. ( -20.30) >consensus GGUGACUUUUUUAAUUGGAAAAUGUUG_AGUUACCCCCGUGUUGAUGGGCCAACAUUUUACCAGUUAAUUUUCGCAUAUUCCAAAACAAAAG .((((.....(((((((((((((((((........(((((....))))).))))))))).))))))))...))))................. (-20.12 = -20.48 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:12 2006