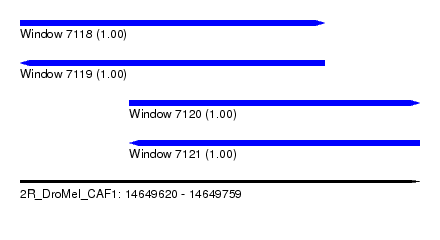
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,649,620 – 14,649,759 |
| Length | 139 |
| Max. P | 1.000000 |

| Location | 14,649,620 – 14,649,726 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 67.54 |
| Mean single sequence MFE | -27.31 |
| Consensus MFE | -17.54 |
| Energy contribution | -16.66 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.64 |
| SVM decision value | 7.47 |
| SVM RNA-class probability | 1.000000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14649620 106 + 20766785 GUUUGCAAAUCAAACUCAUGCAUAGCUUGAGCGAACAAAGCUUAAUAG-CCCUUGCAGAAUAACUACACAUUCAUUUUAAGAAGCCC-----------AUUGUAUUCUGCAAGGGCAA (((((((...........)))).)))((((((.......))))))..(-(((((((((((((........(((.......)))....-----------....)))))))))))))).. ( -34.89) >DroEre_CAF1 4394 101 + 1 GGUUAUAAAUCGAAAUCAUGGCAAGCUU------AUAA----------ACCCUUGUAGAAUAACUA-ACAUUAACAUUAAGUAGCCCGUAGUGCUACAAAAGUAUUCUGCAAGGGCAA .(((((...........)))))......------....----------.(((((((((((((.((.-...(((....)))(((((.......)))))...)))))))))))))))... ( -24.10) >DroYak_CAF1 4381 106 + 1 GGUUGUAAACCGAAAUCAUGCACAGUUUCAGCAAACAA----------ACUCUUGCAGAAUGACUA-ACAUUAACAUUAAGUAGCUUAUAG-ACCAUAAAAGUAUUCUGCAAGAGCAA (((.....)))(((((........))))).........----------.(((((((((((((.((.-..........((((...))))...-........)))))))))))))))... ( -22.95) >consensus GGUUGUAAAUCGAAAUCAUGCAAAGCUU_AGC_AACAA__________ACCCUUGCAGAAUAACUA_ACAUUAACAUUAAGUAGCCC_UAG__C_A_AAAAGUAUUCUGCAAGGGCAA .................................................(((((((((((((.(.....................................))))))))))))))... (-17.54 = -16.66 + -0.88)


| Location | 14,649,620 – 14,649,726 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 67.54 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -16.55 |
| Energy contribution | -16.67 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.53 |
| SVM decision value | 5.49 |
| SVM RNA-class probability | 0.999988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14649620 106 - 20766785 UUGCCCUUGCAGAAUACAAU-----------GGGCUUCUUAAAAUGAAUGUGUAGUUAUUCUGCAAGGG-CUAUUAAGCUUUGUUCGCUCAAGCUAUGCAUGAGUUUGAUUUGCAAAC ..((((((((((((((...(-----------(.(((((.......))).)).))..)))))))))))))-).((((((((((((..((....))...))).)))))))))........ ( -37.80) >DroEre_CAF1 4394 101 - 1 UUGCCCUUGCAGAAUACUUUUGUAGCACUACGGGCUACUUAAUGUUAAUGU-UAGUUAUUCUACAAGGGU----------UUAU------AAGCUUGCCAUGAUUUCGAUUUAUAACC ..(((((((.((((((.....(((((.(....))))))(((((......))-))).)))))).)))))))----------((((------(((.(((.........)))))))))).. ( -25.10) >DroYak_CAF1 4381 106 - 1 UUGCUCUUGCAGAAUACUUUUAUGGU-CUAUAAGCUACUUAAUGUUAAUGU-UAGUCAUUCUGCAAGAGU----------UUGUUUGCUGAAACUGUGCAUGAUUUCGGUUUACAACC ..((((((((((((((((..(((...-...((((...))))......))).-.))).)))))))))))))----------((((..((((((((.......).)))))))..)))).. ( -31.60) >consensus UUGCCCUUGCAGAAUACUUUU_U_G__CUA_GGGCUACUUAAUGUUAAUGU_UAGUUAUUCUGCAAGGGU__________UUGUU_GCU_AAGCUAUGCAUGAUUUCGAUUUACAACC ..((((((((((((((((.............(((...))).............))).)))))))))))))................................................ (-16.55 = -16.67 + 0.12)

| Location | 14,649,658 – 14,649,759 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 67.01 |
| Mean single sequence MFE | -28.39 |
| Consensus MFE | -19.59 |
| Energy contribution | -19.99 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.69 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
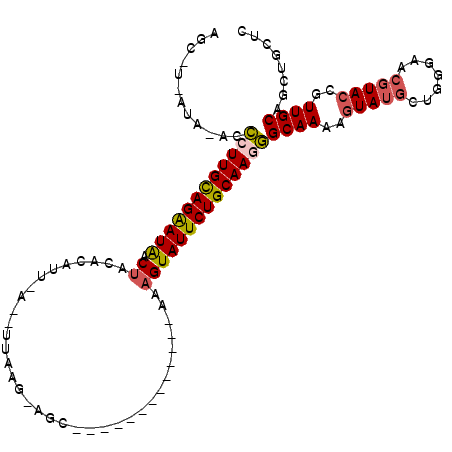
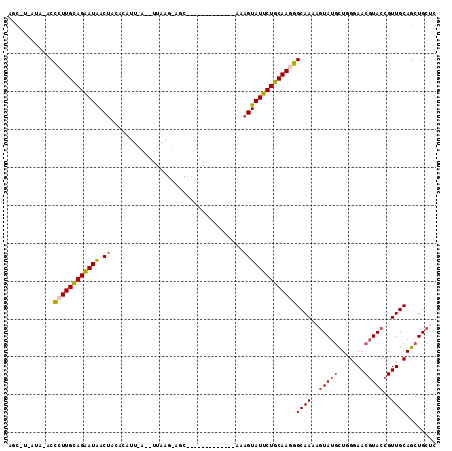
>2R_DroMel_CAF1 14649658 101 + 20766785 AGCUUAAUAG-CCCUUGCAGAAUAACUACACAUUCAUUUUAAGAAGCCC-----------AUUGUAUUCUGCAAGGGCAAAAGUAUUCUGGUAACGUACCGUUGCAGCUGCUC ((((.....(-(((((((((((((........(((.......)))....-----------....))))))))))))))............((((((...)))))))))).... ( -35.09) >DroSec_CAF1 4420 83 + 1 AGCCUGAUAAUCCCUUGCAGAAUAACUACA------------------------------ACAGUAUUCUGCAAAGGCAAUCGUAUGUUGGGAACGUACCGUUGCAGCUGCUC (((.........(.((((((((((.((...------------------------------..)))))))))))).)(((((.((((((.....)))))).))))).))).... ( -25.70) >DroSim_CAF1 4277 83 + 1 AGCCUAAUAAUCCCUUGCAGGAUAACUACA------------------------------AAAGUAUUCUGCAAAGGCAAUCGUAUGUUGGGAACGUACCGUUGCAGCUGCUC (((.........(.((((((((((.((...------------------------------..)))))))))))).)(((((.((((((.....)))))).))))).))).... ( -25.40) >DroEre_CAF1 4426 102 + 1 ----------ACCCUUGUAGAAUAACUA-ACAUUAACAUUAAGUAGCCCGUAGUGCUACAAAAGUAUUCUGCAAGGGCAAAAGUAUGCUAGGAACUUACCGUUGCAGCUGCUC ----------.(((((((((((((.((.-...(((....)))(((((.......)))))...)))))))))))))))....((((.(((.(.(((.....))).)))))))). ( -31.10) >DroYak_CAF1 4419 101 + 1 ----------ACUCUUGCAGAAUGACUA-ACAUUAACAUUAAGUAGCUUAUAG-ACCAUAAAAGUAUUCUGCAAGAGCAAAAAUAUGCUAGGAACGUACCGUUGCAGCUGCUC ----------.(((((((((((((.((.-..........((((...))))...-........))))))))))))))).........(((.(.((((...)))).))))..... ( -24.65) >consensus AGC_U_AUA_ACCCUUGCAGAAUAACUACACAUU_A__UUAAG_AGC_____________AAAGUAUUCUGCAAGGGCAAAAGUAUGCUGGGAACGUACCGUUGCAGCUGCUC ............((((((((((((.((...................................))))))))))))))((((..(((((.......)))))..))))........ (-19.59 = -19.99 + 0.40)

| Location | 14,649,658 – 14,649,759 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 67.01 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -18.37 |
| Energy contribution | -18.21 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
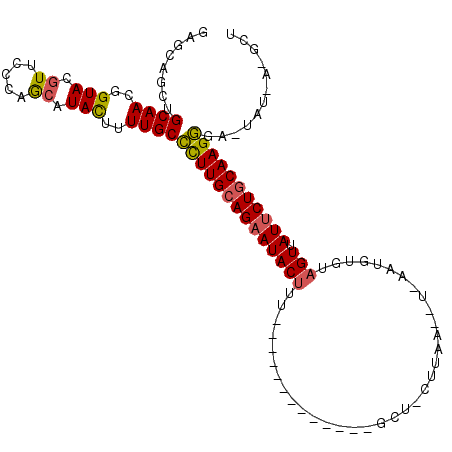
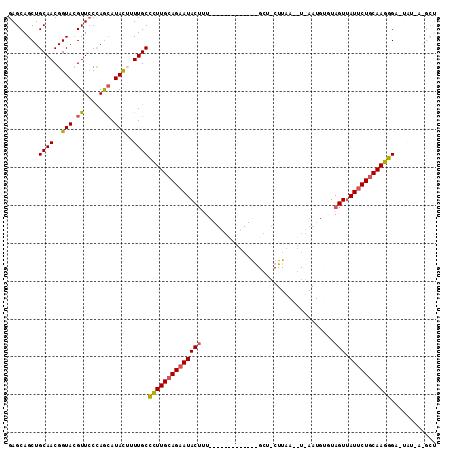
>2R_DroMel_CAF1 14649658 101 - 20766785 GAGCAGCUGCAACGGUACGUUACCAGAAUACUUUUGCCCUUGCAGAAUACAAU-----------GGGCUUCUUAAAAUGAAUGUGUAGUUAUUCUGCAAGGG-CUAUUAAGCU ....((((.....((((...))))...........((((((((((((((...(-----------(.(((((.......))).)).))..)))))))))))))-).....)))) ( -34.50) >DroSec_CAF1 4420 83 - 1 GAGCAGCUGCAACGGUACGUUCCCAACAUACGAUUGCCUUUGCAGAAUACUGU------------------------------UGUAGUUAUUCUGCAAGGGAUUAUCAGGCU ....((((......(((.((.....)).)))(((..((((((((((((((((.------------------------------..))).)))))))))))))...))).)))) ( -26.20) >DroSim_CAF1 4277 83 - 1 GAGCAGCUGCAACGGUACGUUCCCAACAUACGAUUGCCUUUGCAGAAUACUUU------------------------------UGUAGUUAUCCUGCAAGGGAUUAUUAGGCU .(((..((((((.(((((((.........)))..)))).))))))....((((------------------------------(((((.....)))))))))........))) ( -24.60) >DroEre_CAF1 4426 102 - 1 GAGCAGCUGCAACGGUAAGUUCCUAGCAUACUUUUGCCCUUGCAGAAUACUUUUGUAGCACUACGGGCUACUUAAUGUUAAUGU-UAGUUAUUCUACAAGGGU---------- .....((((.(((.....))).).)))........(((((((.((((((.....(((((.(....))))))(((((......))-))).)))))).)))))))---------- ( -28.60) >DroYak_CAF1 4419 101 - 1 GAGCAGCUGCAACGGUACGUUCCUAGCAUAUUUUUGCUCUUGCAGAAUACUUUUAUGGU-CUAUAAGCUACUUAAUGUUAAUGU-UAGUCAUUCUGCAAGAGU---------- .....((((.((((...)))).).)))........((((((((((((((((..(((...-...((((...))))......))).-.))).)))))))))))))---------- ( -26.90) >consensus GAGCAGCUGCAACGGUACGUUCCCAGCAUACUUUUGCCCUUGCAGAAUACUUU_____________GCU_CUUAA__U_AAUGUGUAGUUAUUCUGCAAGGGA_UAU_A_GCU ........((((..(((.((.....)).)))..))))((((((((((((((...................................))).)))))))))))............ (-18.37 = -18.21 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:05 2006