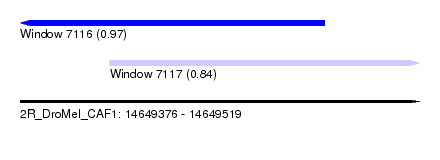
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,649,376 – 14,649,519 |
| Length | 143 |
| Max. P | 0.965901 |

| Location | 14,649,376 – 14,649,485 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.30 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -24.28 |
| Energy contribution | -24.17 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14649376 109 - 20766785 UCCAACUGGAGCCGCAGACUCGUGGGCGCUCAGCCUCGGCUGGUGCGCAGUUCAAAAACGAUUUGGGCUACAUGAAGAAGAUCUUCAAGUAGCG----CUUAA---AAGACACUCU .....((.((((..((((.((((..(((((((((....))))).)))).........)))))))).(((((.(((((.....))))).))))))----)))..---.))....... ( -38.30) >DroPse_CAF1 7631 108 - 1 --CUCCUAGAGCCGCAGCCCCGUGGCCGCUCUGCUUCGGCGGGGGCUCAGUUCAAGAACGACCUCAGCUACAUGAAGAAGAUCUUCAAAUAGUC----CUCCCCCACA-CCCUUU- --......(((((((.(((....))).))(((((....)))))))))).(((....))).......((((..(((((.....)))))..)))).----..........-......- ( -32.50) >DroEre_CAF1 4151 108 - 1 UCCAACUGGAGCCGCAGACACGUGGACGAUCAGCCUCGGCUGGUGCGCAGUUCAAAAACGAUUUGGGCUACAUGAAGAAGAUCUUCAAGUAGCG----CUAAG---AAGCCACUC- (((....)))...........((((.(.((((((....))))))((((.(((((((.....)))))))(((.(((((.....))))).))))))----)....---..)))))..- ( -35.60) >DroWil_CAF1 306 109 - 1 AUAAUCUUGAGCCGCUUCCACGCGGACGCUCUGCCUCGGCGGGAGCUCAAUUCAAAAAUGACCUUAGCUACAUGAAGAAAAUCUUCAAGUAGUUUCCUCUCAA---U--AUAUU-- ......((((((.((.(((....))).))(((((....))))).))))))........(((....((((((.(((((.....))))).)))))).....))).---.--.....-- ( -32.10) >DroYak_CAF1 4138 108 - 1 UCCAACUGGAGCCGCAGACACGUGGGCGAUCCGCCUCGGCUGGUGCGCAGUUCAAAAACGAUUUGGGCUACAUGAAGAAGAUCUUCAAGUAGCG----CUGAA---UAGCCAUUU- (((....)))..(((......)))((((...))))..(((((..((((.(((((((.....)))))))(((.(((((.....))))).))))))----)....---)))))....- ( -36.10) >DroPer_CAF1 7837 108 - 1 --CUCCUAGAGCCGCAGCCCCGUGGCCGCUCUGCUUCGGCGGGGGCUCAGUUCAAGAACGACCUCAGCUACAUGAAGAAGAUCUUCAAAUAGUC----CUCCCCCACA-CCCUUU- --......(((((((.(((....))).))(((((....)))))))))).(((....))).......((((..(((((.....)))))..)))).----..........-......- ( -32.50) >consensus UCCAACUGGAGCCGCAGACACGUGGACGCUCUGCCUCGGCGGGUGCGCAGUUCAAAAACGACCUGAGCUACAUGAAGAAGAUCUUCAAGUAGCG____CUCAA___AA_CCAUUU_ ........(((((((......)))).).((((((....)))))).......)).............(((((.(((((.....))))).)))))....................... (-24.28 = -24.17 + -0.11)

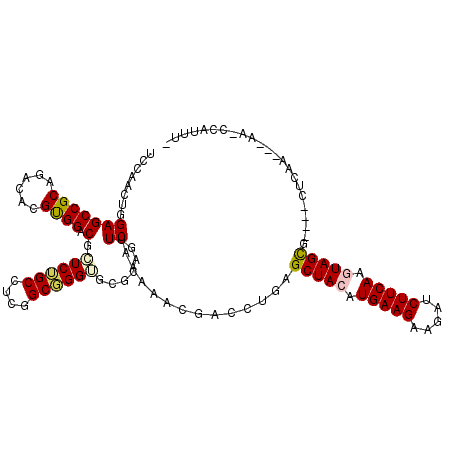
| Location | 14,649,408 – 14,649,519 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.80 |
| Mean single sequence MFE | -47.12 |
| Consensus MFE | -33.99 |
| Energy contribution | -34.50 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14649408 111 + 20766785 CUUCAUGUAGCCCAAAUCGUUUUUGAACUGCGCACCAGCCGAGGCUGAGCGCCCACGAGUCUGCGGCUCCAGUUGGAGUUGCGAGUGGGGAAAGGACUGGGGUUGGUGUUU ......((((..((((.....))))..))))(((((((((..((((...(.(((((.....(((((((((....))))))))).))))))...)).))..))))))))).. ( -46.90) >DroSec_CAF1 4180 111 + 1 CUUCAUGUAGCCCAAAUCGUUUUUGAACUGCGCACCAGCCGAGGCUGAUCGCCCACGAGUUUGCGGCUCCAGUUGGAGUUGCGAGUGGGGAAAGGACUGGGGUUGGUGCUU ......((((..((((.....))))..))))(((((((((..((((..((.(((((....((((((((((....)))))))))))))))))..)).))..))))))))).. ( -51.40) >DroEre_CAF1 4182 111 + 1 CUUCAUGUAGCCCAAAUCGUUUUUGAACUGCGCACCAGCCGAGGCUGAUCGUCCACGUGUCUGCGGCUCCAGUUGGAGUUGCGAGUGGGGAAAGGACUGGGGUUGGUGCUU ......((((..((((.....))))..))))(((((((((..((((..((.(((((.....(((((((((....))))))))).)))))))..)).))..))))))))).. ( -47.60) >DroYak_CAF1 4169 111 + 1 CUUCAUGUAGCCCAAAUCGUUUUUGAACUGCGCACCAGCCGAGGCGGAUCGCCCACGUGUCUGCGGCUCCAGUUGGAGCUGCGAGUGGGGAAAGGACUGAGGUUGGUGCUU ......((((..((((.....))))..))))(((((((((..(((...((.(((((.....(((((((((....))))))))).)))))))...).))..))))))))).. ( -50.70) >DroMoj_CAF1 339 108 + 1 CUUCAUAUAGCUAAGAUCGUUUUUGAGCUGCGCCCCCGCCGAGGCAGAGCGACUGCGCUGCUGCGGCUCAAAGGC-GGGCGCGGGCGGUGCCAGG--UGCGAACUGUGCCC ..................(((((((((((((((....))...(((((.((....)).))))))))))))))))))-((((((((....(((....--.)))..)))))))) ( -46.60) >DroAna_CAF1 7532 111 + 1 UUUCAUGUAGCUUAUGUCGUUUUUGAACUGCGCCCCAGCUGAAGCGGAACGUCCACGCGCUUCUGGCUCCAGUUGGAGCUGAGAGUGUGGAAAGGAUUGGGGAUGGUGCUU ..((((((((.(((.........))).)))).(((((((....)).....((((.(((((((..((((((....))))))..)))))))....)))))))))))))..... ( -39.50) >consensus CUUCAUGUAGCCCAAAUCGUUUUUGAACUGCGCACCAGCCGAGGCUGAUCGCCCACGAGUCUGCGGCUCCAGUUGGAGUUGCGAGUGGGGAAAGGACUGGGGUUGGUGCUU ......((((..((((.....))))..))))(((((((((..(((...((.(((((.....(((((((((....))))))))).)))))))...).))..))))))))).. (-33.99 = -34.50 + 0.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:02 2006