| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,649,073 – 14,649,178 |
| Length | 105 |
| Max. P | 0.575696 |

| Location | 14,649,073 – 14,649,178 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
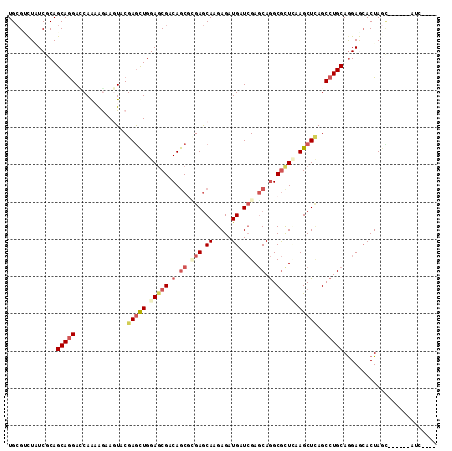
| Mean pairwise identity | 77.97 |
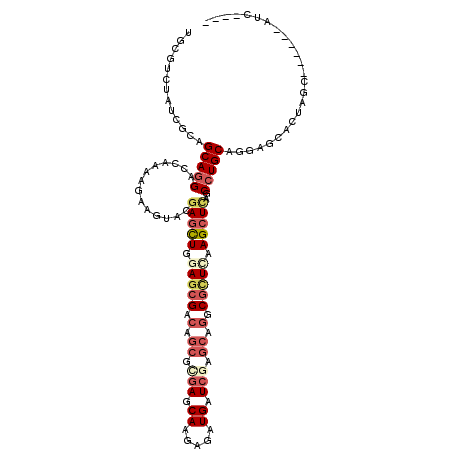
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -19.85 |
| Energy contribution | -21.49 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14649073 105 - 20766785 UGCGACUGUCACAGCAAGAGCAGAGAAAGUUCGAACUGGAGCGGCAGCAGGAGCAGGAGAUGAUCGAGCAGGCGUUAAAGCUUAGCCUGCAAGAGCACUAAU------AUA---- (((..((((..(((...((((.......))))...)))..))))..)))((.((..((.....))..((((((...........))))))....)).))...------...---- ( -31.20) >DroVir_CAF1 17 114 - 1 UGCGUCUUUCGCAGCAGGAUCAGCAAAAAUACGAGCUGGAGCGCCAGCGUGAGCAAGAGAUGAUCGAGCAGGCGCUUAAGCUCAGCCUGCAGGAGCACUAGCCAUCCUAC-GAAC ((((.....))))(((((....(........)(((((.(((((((.((....))..((.....)).....))))))).)))))..)))))((((((....))..))))..-.... ( -43.50) >DroPse_CAF1 7328 111 - 1 UGCGUUUAUCGCAGCAGGACCAAAAGAAGUACGAGCUGGAGCGACAGCGCGAGCAAGAGAUGAUCGAGCAGGCGCUCAAGCUCAGCCUGCAGGAACACUAGCCAAUCAAUC---- ((((.....))))(((((((........))..(((((.(((((...((.(((.((.....)).))).))...))))).)))))..))))).((........))........---- ( -37.00) >DroMoj_CAF1 1 101 - 1 UGCGCCUCUCCCAGCAGGAGCAGCAGAAGUACGAGCUGGAGCGACAGCGCGAGCAGGAGAUGAUCGAGCAGGCGCUCAAGCUCAGCCUGCAGGAGAACUAG-------------- ......(((((..(((((....((....))..(((((.(((((...((.(((.((.....)).))).))...))))).)))))..))))).))))).....-------------- ( -40.10) >DroAna_CAF1 7207 105 - 1 UGAGACUGUCACAGCAGGAUCAAAGGAAAUACGAGUUGGAACGGCAACAGGAACAGGAGAUGAUAGAUCAAGCACUGAAGCUGAGCCUGCAGGAGCACUAAU------GUC---- (((..((((....))))..)))............(((((...(((..(((...(((....(((....)))....)))...))).)))(((....))))))))------...---- ( -21.00) >DroPer_CAF1 7534 111 - 1 UGCGUUUAUCGCAGCAGGACCAAAAGAAGUACGAGCUGGAGCGACAGCGCGAGCAAGAGAUGAUCGAGCAGGCGCUCAAGCUCAGCCUGCAGGAACACUAGCCAAUCAAUC---- ((((.....))))(((((((........))..(((((.(((((...((.(((.((.....)).))).))...))))).)))))..))))).((........))........---- ( -37.00) >consensus UGCGUCUAUCGCAGCAGGACCAAAAGAAGUACGAGCUGGAGCGACAGCGCGAGCAAGAGAUGAUCGAGCAGGCGCUCAAGCUCAGCCUGCAGGAGCACUAGC______AUC____ .............(((((..............(((((.(((((.(.((.(((.((.....)).))).)).).))))).)))))..)))))......................... (-19.85 = -21.49 + 1.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:59 2006