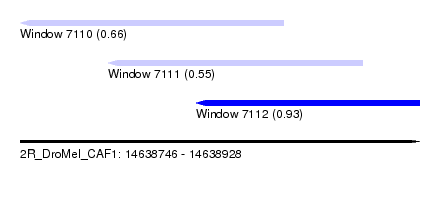
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,638,746 – 14,638,928 |
| Length | 182 |
| Max. P | 0.930357 |

| Location | 14,638,746 – 14,638,866 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.78 |
| Mean single sequence MFE | -43.75 |
| Consensus MFE | -20.94 |
| Energy contribution | -18.78 |
| Covariance contribution | -2.16 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
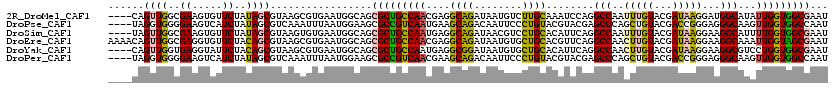
>2R_DroMel_CAF1 14638746 120 - 20766785 GCAGCGCUGCCAACGAGGCAGAUAAUGUCUUGCAAAUCCAGGCCAAUUUGUACGAUAAGGAUGGCAUAUUGGUGGCGAAUGCCACGUCGGACCAAAAACUUGGCGGCAAGCUGCAAGUGA ((((((((((((((((((((.....)))))))....(((..((((.(((........))).))))......(((((....)))))...)))........))))))))..)))))...... ( -46.60) >DroVir_CAF1 1531 120 - 1 GCACAGCCGCUAAUGAAGCGGACAAUACCCUGUACGCACGGCUGCAGCUGUACAGCAGGGAGGGCAUACUAGUGGCCAAUGCCACCUCGGACAGCAAGCUCUUGGGUUCAUUGAAAGUUG ((...((((((.....))))).)....((((((..(.(((((....))))).).))))))...))......(((((....)))))...((((..(((....))).))))........... ( -37.80) >DroPse_CAF1 1511 120 - 1 GAAGCGCCGUCAAUGAAGCAGACAAUUCCCUGUACGUACGAGCCCAGCUGUACGACCGGGAGGGCAAGUUGGUGGCCAAUGGCACCUCGGAUGAGGAACUUUGUGGAAGUCUGCAGGUGG ....((((.((...)).((((((..(((((.((.(((((.(((...)))))))))).))))).(((((((....(((...))).((((....))))))).))))....)))))).)))). ( -42.50) >DroEre_CAF1 1439 120 - 1 GCAGCGCUGCCAACGAGGCAGAUAAUGUGCUGCACGUUCAGGCCAACUUGUACGAUAAGGAAGGCAAAUUGGUAGCGAAUGCCACGUCGGACGGAAAACUAGGCGGCAAGUUGCAAGUGA (((((.(((((.....)))))......((((((.(((((..(((..(((........)))..)))....(((((.....)))))....))))).........)))))).)))))...... ( -41.10) >DroYak_CAF1 1405 120 - 1 GCAGCGCUGCCAAUGAGGCGGAUAAUGUGCUGCACAUUCAGGCCAACUUGUACGAUAAGGAAGGCGUCCUGGUGGCGAAUGCCACAUCGGACGGCAAACUAGGCGGCAAGUUGCAGGUGA (((((((((((.....))))......))))))).((((...((.(((((((.((...((...(.(((((..(((((....)))))...))))).)...))...))))))))))).)))). ( -48.80) >DroPer_CAF1 1511 120 - 1 GAAGCGCCGUCAACGAAGCAGACAAUUCCCUGUACGUACGAGCCCAGCUGUACGACCGGGAGGGCAAGUUGGUGGCCAAUGGCACCUCGGAUGAGGAGCUUUGUGGAAGUCUGCAGGUGG ....(((((....)...((((((..(((((.((.(((((.(((...)))))))))).))))).............(((..(((.((((....)))).)))...)))..)))))).)))). ( -45.70) >consensus GCAGCGCCGCCAACGAAGCAGACAAUGCCCUGCACGUACAGGCCAAGCUGUACGACAAGGAGGGCAAAUUGGUGGCCAAUGCCACCUCGGACGAAAAACUUGGCGGCAAGCUGCAAGUGA .....(((((((.....((((........))))........((((..((........))..))))....)))))))..........................((((....))))...... (-20.94 = -18.78 + -2.16)

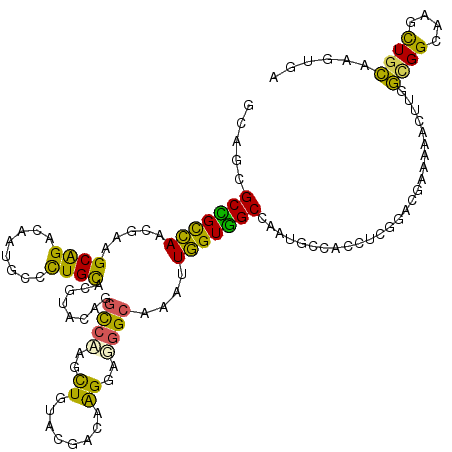
| Location | 14,638,786 – 14,638,902 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -38.37 |
| Consensus MFE | -21.41 |
| Energy contribution | -20.83 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14638786 116 - 20766785 ----CAGUUGGCGAAGUGUUCUAUAGCGUAAGCGUGAAUGGCAGCGCUGCCAACGAGGCAGAUAAUGUCUUGCAAAUCCAGGCCAAUUUGUACGAUAAGGAUGGCAUAUUGGUGGCGAAU ----..((((((..((((((((((.((....))....)))).))))))))))))((((((.....))))))((....(((.((((.(((........))).))))....)))..)).... ( -37.40) >DroPse_CAF1 1551 116 - 1 ----UAGGUGGGGAAGUCAUCUAUAGCGUCAAAUUUAAUGGAAGCGCCGUCAAUGAAGCAGACAAUUCCCUGUACGUACGAGCCCAGCUGUACGACCGGGAGGGCAAGUUGGUGGCCAAU ----(((((((.....)))))))..((.((..........)).))(((..((((...((......(((((.((.(((((.(((...)))))))))).))))).))..))))..))).... ( -35.30) >DroSim_CAF1 1481 116 - 1 ----UAGUUGGCGAAGUGUUCUAUAGCGUAAGUGUGAAUGGCAGCGCUGCCAAUGAGGCAGAUAACGUCCUGCACAUUCAGGCCAAUUUGUACGAUAAGGAAGGCAUUUUGGUGGCGAAU ----..((((.(((((((((((.((.((((.((.(((((((((((((((((.....)))))....))..)))).)))))).)).......)))).)).)))..)))))))).)))).... ( -37.40) >DroEre_CAF1 1479 120 - 1 AAAACAGUUGGCGAGGUGUUCUACAGCGUAAGCGUGAAUGGCAGCGCUGCCAACGAGGCAGAUAAUGUGCUGCACGUUCAGGCCAACUUGUACGAUAAGGAAGGCAAAUUGGUAGCGAAU ......((((.(((.((.((((....((((.((.(((((((((((((((((.....))))......))))))).)))))).)).......))))....)))).))...))).)))).... ( -42.70) >DroYak_CAF1 1445 116 - 1 ----CAGUUGGUGAGGUAUUCUACAGCGUAAGCGUGAAUGGCAGCGCUGCCAAUGAGGCGGAUAAUGUGCUGCACAUUCAGGCCAACUUGUACGAUAAGGAAGGCGUCCUGGUGGCGAAU ----..((((((.............((....)).(((((((((((((((((.....))))......))))))).)))))).))))))((((.((...((((.....))))..)))))).. ( -40.60) >DroPer_CAF1 1551 116 - 1 ----UAGGUGGGGAAGUCAUCUAUAGCGUCAAAUUUAAUGGAAGCGCCGUCAACGAAGCAGACAAUUCCCUGUACGUACGAGCCCAGCUGUACGACCGGGAGGGCAAGUUGGUGGCCAAU ----(((((((.....)))))))..((.((..........)).))(((..((((...((......(((((.((.(((((.(((...)))))))))).))))).))..))))..))).... ( -36.80) >consensus ____CAGUUGGCGAAGUGUUCUAUAGCGUAAGCGUGAAUGGCAGCGCUGCCAACGAGGCAGAUAAUGUCCUGCACAUUCAGGCCAACUUGUACGAUAAGGAAGGCAAAUUGGUGGCGAAU ......((((..((.....))..)))).................(((((((((....((((........))))........(((..(((((...)))))...)))...)))))))))... (-21.41 = -20.83 + -0.58)
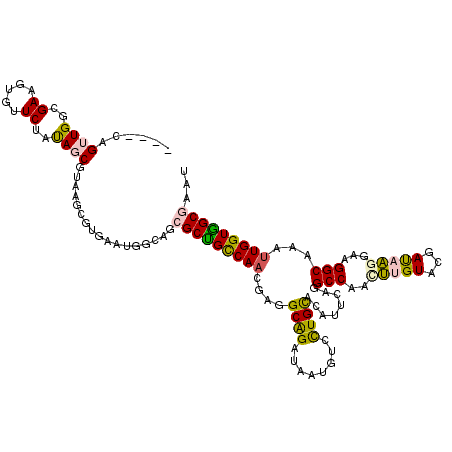
| Location | 14,638,826 – 14,638,928 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.76 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -18.29 |
| Energy contribution | -16.94 |
| Covariance contribution | -1.35 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14638826 102 - 20766785 UAACUUAUUUAAUA-CUGU--AAUACUUU----------CAGUUGGCGAAGUGUUCUAUAGCGUAAGCGUGAAUGGCAGCGCUGCCAACGAGGCAGAUAAUGUCUUGCAAAUCCA ..............-.(((--........----------..((((((..((((((((((.((....))....)))).))))))))))))((((((.....)))))))))...... ( -27.80) >DroVir_CAF1 1611 102 - 1 GAGCUAU---AAUAUCUGCAGUUCAUUUU----------CAGUGGGCACUGUGAAUUACAGCAUUGCGGUGAAUGGCACAGCCGCUAAUGAAGCGGACAAUACCCUGUACGCACG ..(((((---.....(((((((((((...----------..)))))).(((((...)))))...)))))...)))))(((((((((.....)))))........))))....... ( -26.40) >DroSec_CAF1 1638 102 - 1 UAACUUAUUUAAAA-CUGU--AAUGCUUU----------UAGUUGGCGAAGUGUUCUAUAGCGUAAGCGUGAAUGGCAGCGCUGCCAAUGAGGCAGAUAAUGUCCUCCACAUCCA ..............-.(((--........----------..((((((..((((((((((.((....))....)))).))))))))))))((((((.....)).)))).))).... ( -25.50) >DroSim_CAF1 1521 102 - 1 UAACUUAAUUAAAA-CUGU--AAUACUUU----------UAGUUGGCGAAGUGUUCUAUAGCGUAAGUGUGAAUGGCAGCGCUGCCAAUGAGGCAGAUAACGUCCUGCACAUUCA ..(((((.......-((((--((((((((----------........)))))))..)))))..))))).(((((((((((((((((.....)))))....))..)))).)))))) ( -26.10) >DroEre_CAF1 1519 109 - 1 UAAUUU---UAAUA-CUGG--AAUACUUGCCCUUAAAAACAGUUGGCGAGGUGUUCUACAGCGUAAGCGUGAAUGGCAGCGCUGCCAACGAGGCAGAUAAUGUGCUGCACGUUCA ......---.....-.(((--((((((((((.............))).))))))))))..((....)).(((((((((((((((((.....))))......))))))).)))))) ( -38.82) >DroYak_CAF1 1485 102 - 1 UAAUAAUAAUAAUA-UUGU--AAUACUUU----------CAGUUGGUGAGGUAUUCUACAGCGUAAGCGUGAAUGGCAGCGCUGCCAAUGAGGCGGAUAAUGUGCUGCACAUUCA ..............-.(((--((((((((----------........)))))))..))))((....)).(((((((((((((((((.....))))......))))))).)))))) ( -31.00) >consensus UAACUUA_UUAAUA_CUGU__AAUACUUU__________CAGUUGGCGAAGUGUUCUACAGCGUAAGCGUGAAUGGCAGCGCUGCCAAUGAGGCAGAUAAUGUCCUGCACAUUCA ...............(((...((((((((..................))))))))...))).......(((...((((...(((((.....)))))....))))...)))..... (-18.29 = -16.94 + -1.35)
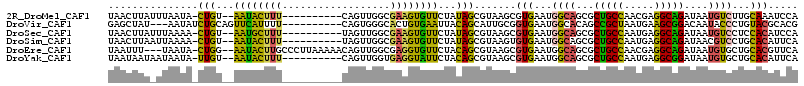
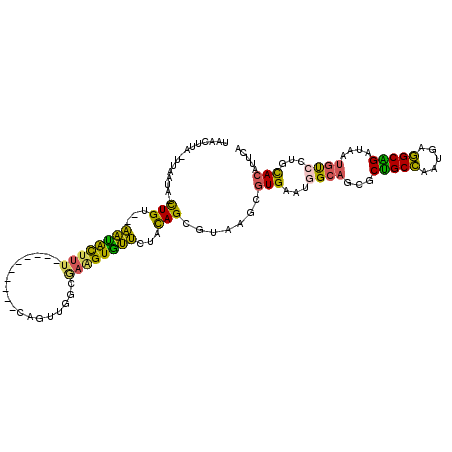

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:55 2006